Abstract
The mitochondrial genome sequence of Pseudoxenodon stejnegeri (Squamata: Colubridae: Pseudoxenodontinae) from Taishun County, Zhejiang Province, China, which is 18,475 bp in length and contains 25 tRNAs (including extra two tRNA-Tyr genes and extra one tRNA-Met gene), two rRNAs, 13 protein-coding genes and two identical control regions. The overall AT content of the mitogenome is 59.6% (A = 32.6%, T = 27%, C = 27%, G = 13.4%). In BI and ML phylogenetic analyses, the monophyly of the family Colubridae was well supported and P. stejnegeri was a basal clade of Colubridae.
The taxonomic position of Genus Pseudoxenodon (Squamata: Colubridae: Pseudoxenodontinae) remains controversial. Species of Pseudoxenodon are widely distributed throughout southern and southeastern Asia. Pseudoxenodon stejnegeri is widely distributed in southern China (Zhang and Huang Citation2013). In this study, we sequenced the complete mitochondrial genome of Pseudoxenodon stejnegeri and combined with the existing mitochondrial genome sequence of Colubroidea to discuss the relationship of Genus Pseudoxenodon.
The sample of P. stejnegeri (ZJTS20180528) was collected from Taishun (N 119.68°, E 27.71°), Zhejiang Province, China. The sample was identified and stored at −20 °C in College of Life and Environmental Science, Wenzhou University, China. Total genomic DNA was extracted from muscle tissue using Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China) and stored in the laboratory. In this study, the complete mitochondrial genome was obtained according to the modified universal primers (Liao et al. Citation2020; Zong et al. Citation2020). Subsequently, the remaining gaps were sequenced by designing species-specific primers according to previously obtained sequences. All PCR products were sequenced in both directions by the Sangon Biotech Company (Shanghai, China). The mitochondrial genome was deposited in GenBank with an accession number MW018358.
The complete mitogenome of P. stejnegeri is a circular DNA molecule with a total length of 18,475 bp. It contains 25 tRNAs (including extra two tRNA-Tyr genes and extra one tRNA-Met gene), two rRNAs, 13 protein-coding genes and two identical control regions. Three tRNA-Tyr genes are identical but the extra one tRNA-Met is different. The overall AT content of the whole mitogenome is 59.6%. In the 13 protein-coding genes, ND4 gene is used GTG as the start codon, while the remaining 12 protein-coding genes used ATN (N stands for A, T, C, G) as the start codon. Some protein-coding genes are used TAA as a stop codon. However, ND1 and COXI are used AGG as a stop codon, ND2, ND4 and ND4L used TAG as a stop codon, ND3 and ND6 used AGA as a stop codon, and COXIII, Cytb and ND5 end with an incomplete stop codon (T–).
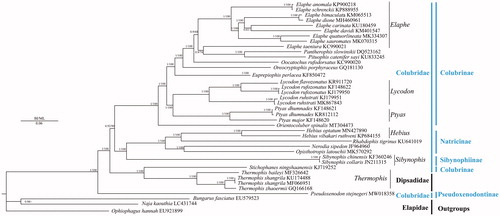
To construct a phylogenetic relationship of P. stejnegeri, 39 mitochondrial genomes of 33 Squamata species including three mitochondrial genomes as outgroups (Bungarus fasciatus, Naja kaouthia, Ophiophagus hannah) were downloaded from GenBank. Phylogenetic relationships were reconstructed using the Bayesian inference (BI) method implemented in MrBayes version 3.1.2 (Huelsenbeck and Ronquist Citation2001) and maximum likelihood (ML) in RAxML 8.2.0 (Stamatakis Citation2014) based on 13 protein-coding genes. To select conserved regions of the putative nucleotide sequences, each alignment was analyzed with Gblocks 0.91 b (Castresana Citation2000) using default settings. In BI and ML phylogenetic analyses (), the monophyly of Colubridae was failed to support because the Family Dipsadidae was clustered into Family Colubridae. The monophyly of Genus Elaphe was failed to support because the clade of Pantherophis slowinskii and Pituphis catenifer sayi was clustered into Genus Elaphe, whereas the monophyly of Genera Hebius, Lycodon, Ptyas, Sibynophis and Thermophis was well support. P. stejnegeri was a sister clade of ((Dipsadidae + Stichophanes ningshaanensis)+((Sibynophiinae + Natricinae)+Colubrinae)).
Figure 1. Phylogenetic tree of the relationships among 34 species of Colubridae including Pseudoxenodon stejnegeri in this study and three outgroups (Bungarus fasciatus, Naja kaouthia, Ophiophagus Hannah) (Singchat et al. Citation2019), were based on the nucleotide dataset of the 13 mitochondrial protein-coding genes. Numbers around the nodes are the posterior probabilities of BI (left) and the bootstrap values of ML (right). The GenBank numbers of all species are shown in the figure.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MW018358].
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Liao J, Tang M, Peng L, Jiang L, You Z, Chen W. 2020. The complete mitochondrial genome sequence of big-eyed Mountain keelback Pseudoxenodon macrops. Mitochondrial DNA B. 5(1):736–737.
- Singchat W, Areesirisuk P, Sillapaprayoon S, Muangmai N, Baicharoen S, Suntrarachun S, Chanhome L, Peyachoknagul S, Srikulnath K. 2019. Complete mitochondrial genome of Siamese cobra (Naja kaouthia) determined using next-generation sequencing. Mitochondrial DNA B. 4(1):577–578.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang B, Huang S. 2013. Relationship of old world Pseudoxenodon and new world Dipsadinae, with comments on underestimation of species diversity of Chinese Pseudoxenodon. Asian Herpetol Res. 4:155–165.
- Zong H, Zhao T, Hu T, Feng D, Li J, Du R, Fu C. 2020. The complete mitochondrial genome of Pseudoxenodon macrops. Mitochondrial DNA B. 5(2):1473–1474.
