Abstract
Laplacea alpestris is a member of the genus Laplacea, which distributes in Central and South America. Genetic information of L. alpestris would provide guidance for the phylogenetic position of this species. Here, we reported and characterized its complete chloroplast (cp) genome using Illumina pair-end sequencing data. The total chloroplast genome size of this species was 157,211 bp, including inverted repeats (IRs) of 26,103 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 86,749 and 18,256 bp, respectively. A total of 132 genes, including 37 tRNA, 8 rRNA, and 87 protein-coding genes were identified. Phylogenetic analysis showed that L. alpestris formed a monophyletic clade with Laplacea fruticosa, and then grouped with Apterosperma oblata. The systematic position of Southeast Asian Laplacea species needs further studies.
Laplacea Kunth, with ca. 30 species, mainly distributed in South and Central America, Malaya, Indonesia (Kobuski Citation1949, Citation1950). The genus was built in 1822 based on the type species (L. speciosa Dyer) from Peru (Humboldt et al. Citation1822). The systematic position of Laplacea changed significantly among different taxonomic treatments based on morphological and floral ontogenic evidence, and even was included in Gordonia s.l. (Airy-Shaw Citation1936; Sealy Citation1958; Keng Citation1962; Ye Citation1990; Tsou Citation1998). Molecular phylogenetic analysis based on rbcL and matK, and the chloroplast genome sequences suggested that only Gordonia brandegeei H. Keng nom. nov. (=Laplacea grandis) (Keng Citation1980) should be retained in Gordonia s.s., other species from Laplacea were members of Theeae (Prince and Parks Citation2001; Yu et al. Citation2017). However, only scarce species were included in previous studies and only one chloroplast genome was reported for Laplacea (Laplacea fruticosa, Yu et al. Citation2017). In this study, we present the complete chloroplast genome sequence of Laplacea alpestris Dyer using Illumina sequencing technology.
Leaf sample of L. alpestris was obtained from the Herbarium of University of Florida (FLAS, voucher FLAS 180,103), the specimen was collected from Massif de la Selle of Haiti. Genomic DNA was isolated using a modified CTAB approach (Doyle and Doyle Citation1987). The 150 bp pair-end reads were sequenced based on the Illumina Hi-Seq 2500 platform. Totally, 14,086,309 reads in size of 4.71 G were obtained for the next analysis. The chloroplast genome was de novo assembled by GetOrganelle script (Jin et al. Citation2020), with SPAdes version 3.10.1 as assembler (Bankevich et al. Citation2012), and visualized the paths of the cp genome using Bandage version 0.8.1 (Wick et al. Citation2015). Geneious version 8.0.2 (Kearse et al. Citation2012) was used to annotate the L. alpestris and then submit to Genebank (the accession number is MT916289). The size of chloroplast genome of L. alpestris is 1,57,211 bp. The GC content of the genome is 37.2%. The length of inverted repeats (IR), large single copy (LSC), and small single copy (SSC) were 26,103, 86,749, and 18,256 bp, respectively. The chloroplast genome of L. alpestris contained 132 genes, with 8 rRNA genes, 37 tRNA genes, and 87 protein-coding genes. Annotation revealed that 4 rRNA genes, 7 tRNA genes, and 7 protein-coding genes were duplicated in the IR region.
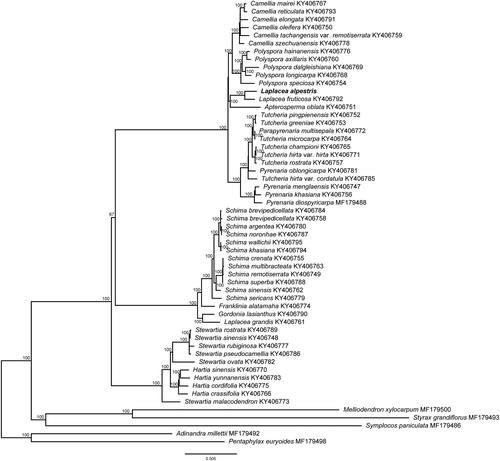
To confirm the phylogenetic position of L. alpestris, we conducted the phylogenetic analysis by combining the chloroplast genome of L. alpestris and other 55 species (including 50 ingroups from Theaceae and 5 outgroups). Sequences were aligned using MAFFT version 7.407 (Katoh and Standley Citation2013) with the Auto algorithm. RAxML (Stamatakis Citation2014) was used to build a maximum likelihood (ML) tree, and bootstrap support (BS) were calculated using 1000 replicates. The maximum likelihood phylogenetic tree revealed that L. alpestris and L. fruticosa formed a monophyletic clade (BS = 100%), which was closely related to Apterosperma oblata (). However, only species of Laplacea from Central and South America were studied till now (i.e. L. alpestris, Laplacea fruticosa, and Laplacea portoricensis) (Prince and Parks Citation2001; Yu et al. Citation2017), whether species from Southeast Asia will fall into Laplacea or Gordonia s.s. need further researches. The complete chloroplast genome of L. alpestris would be useful for the genetic diversity studies of this species and provided new molecular data to illuminate the phylogenetic relationships within Theaceae.
Figure 1. Maximum likelihood tree of Theaceae based on 55 complete chloroplast genome sequences, including Laplacea alpestris (GenBank ID: MT916289) sequenced in this study. The bootstrap support values are shown beside the nodes. Five representative taxa of Styracaceae (Melliodendron xylocarpum, MF179500; Styrax grandiflorus MF179493), Symplocaceae (Symplocos paniculata, MF179486), and Pentaphylacaceae (Adinandra millettii MF179492; Pentaphylax euryoides MF179498) from Ericales were used as outgroups.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT916289.
Additional information
Funding
References
- Airy-Shaw HK. 1936. Notes on the genus Schima and on the classification of the Theaceae-Camellioideae. Kew Bull. 1936(9):496–500.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Humboldt AV, Bonpland A, Kunth CS. 1822. Nova genera et species plantarum. Paris, France: Lutetiae Parisiorum.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Keng H. 1962. Comparative morphological studies in the Theaceae. Univ California Public Bot. 33(4):269–384.
- Keng H. 1980. On the unification of Laplacea and Gordonia (Theaceae). Gardens’ Bull. 33(2):303–311.
- Kobuski CE. 1949. Studies in the Theaceae XVIII. The West Indian species of Laplacea. J Arnold Arboretum. 30:166–186.
- Kobuski CE. 1950. Studies in the Theaceae. XX. Notes on the South and Central American species of Laplacea. J Arnold Arboretum. 31(4):405–429.
- Prince LM, Parks CR. 2001. Phylogenetic relationships of Theaceae inferred from chloroplast DNA sequence data. Am J Bot. 88(12):2309–2320.
- Sealy JR. 1958. A revision of the genus camellia. London: Royal Horticultural Society.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tsou CH. 1998. Early floral development of Camellioideae (Theaceae). Am J Bot. 85(11):1531–1547.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Ye CX. 1990. A discussion on relationship among the genera in Theoideae (Theaceae). Acta Sci Nat Univ Sunyatseni. 29(1):74–81.
- Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ. 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215(3):1235–1248.
