Abstract
Magnolia ofeliae A. Vázquez & Cuevas, a plant species endemic to south Jalisco, Mexico, is a critically endangered (CR) species based on the IUCN Red List. In this study, we assembled its complete chloroplast (cp) genome. The total genome size of M. ofeliae was 159,839 bp including four subregions: a large single-copy (LSC) region of 88,027 bp and a small single-copy (SSC) region of 18,752 bp separated by a pair of identical inverted repeat regions (IRs) of 26,530 bp each. The GC content of the cp genome of M. ofeliae is 39.3%. The cp genome encoded a set of 113 genes, containing 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis results that M. ofeliae is a sister to all other magnolias in the subfamily Magnolioideae.
An endemic species in Mexico, Magnolia ofeliae A. Vázquez & Cuevas is only distributed in the type locality (Vázquez-García et al. Citation2013). Despite intensive explorations in the Talpa de Allende region and elsewhere in Jalisco (Rivers et al. Citation2016), it is extremely rare. A second population (ca. 20 adult trees) was recently found in Santa Gertrudis, 20 km east of the type locality, with nearly 20 trees. Therefore, it is categorized as a critically endangered (CR) species in recently revised the red list of Magnoliaceae (Rivers et al. Citation2016; https://www.iucnredlist.org/species/67513575/67513783). This species is included in section Talauma in the recent classification systems of Magnoliaceae (Figlar and Nooteboom Citation2004, Wang et al. Citation2020) because the plant has circumscissile dehiscing folicetum (Vázquez-García et al. Citation2013).
Section Talauma, despite being the largest group in the family (ca. 100 spp.; Vázquez-García et al. Citation2016), is one of the less well-studied lineages of Magnoliaceae based on the recent classification system (Figlar and Nooteboom Citation2004). Up to date, there are 43 chloroplast (cp) genomes in the Magnoliaceae that have been registered in the organelle genome resources of the GenBank and none of them is from the section Talauma. In this study, we determined a complete sequence of the cp genome of M. ofeliae (GenBank accession no. MT522025), as a representative of the sect. Talauma in Magnoliaceae.
Leaf samples were collected from a tree in the Vallarta Botanica Garden (N20°27′55.14′′, W105°17′31.21′′), one of the ex situ conservation stations of Mexican magnolias. The tree was grown from seeds of the type locality, collected in June 2011 (Vázquez-García 1968, IBUG). A voucher specimen is deposited in the herbarium of the Sungshin University, Korea (S. Kim 2019-0071, SWU).
DNA extraction was performed using a commercial plant DNA extraction kit (ExgeneTM; GeneAll, Seoul, Korea). The extracted DNA was sequenced using the Illumina platform at Macrogen Co. (Seoul, Korea). Paired-end sequencing generated a total of 21,451,116 raw reads after removing adapter sequences. The raw data is deposited in the GenBank SRA database (PRJNA666275). There was no need for the filtering of low-quality sequences because the Phred quality score of most of the reads was over 40 by eye examination of raw reads in Geneious Prime version 9.0.5 (Kearse et al. Citation2012).
The raw reads of M. ofeliae were mapped against reference sequences, multiple cp genomes previously published [M. sinica (NC_023241), M. grandiflora (NC_020318), and M. tripetala (NC_024027)], using Geneious Prime version 9.0.5 (Kearse et al. Citation2012). Border sequences of two inverted repeats and sequences of low coverage parts were checked manually, and the mapping for ambiguous regions is repeated. To verify the sequence, we tried de novo assembly using mapped reads on the reference using Geneious Prime version 9.0.5 (Kearse et al. Citation2012) with ‘medium sensitivity’ option, and the assembled result was exactly the same as mapped consensus. The completed sequence was annotated with the previously reported genome of M. kobus (GenBank Accession No. NC_023237) using Geneious Prime version 9.0.5 (Kearse et al. Citation2012).
The size of the cp genome of M. ofeliae is 159,839 bp, including four subregions: a large single-copy (LSC) region of 88,027 bp and a small single-copy (SSC) region of 18,752 bp separated by a pair of identical inverted repeat regions (IRs) of 26,530 bp each. The GC content of the cp genome of M. ofeliae is 39.3%. The cp genome encoded a set of 113 genes, containing 79 protein-coding genes, 30 tRNAs, and four rRNAs.
Phylogenetic analysis was performed using the cp genome sequence of M. ofeliae and 18 cp genomes that are from representatives of subfamily Magnolioideae and two outgroups (L. chinense and L. tulipifera) from subfamily Liriodendroideae (). The alignment of these sequences was performed by the MAFFT module in Geneious Prime version 9.0.5 (Kearse et al. Citation2012). A maximum likelihood analysis has performed using IQ-TREE (Nguyen et al. Citation2015) with the best model (TVM + F + I) including 1000 replication of bootstrap analyses. The result showed that M. ofeliae, as a member of sect. Talauma, is a sister to all other magnolias in the subfamily Magnolioideae and the relationship is highly supported.
Figure 1. A maximum-likelihood tree based on an alignment of entire cp genome sequences that are from M. ofeliae and selected members of Magnoliaceae.
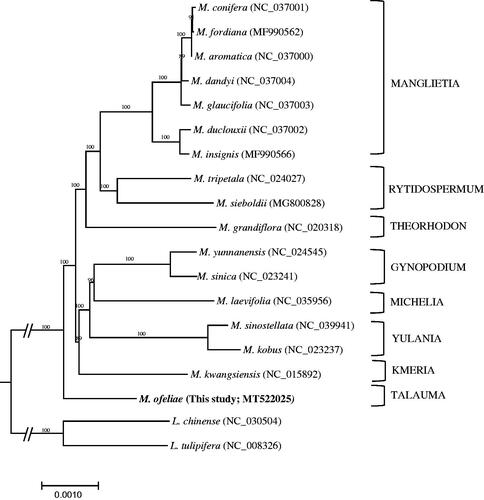
The cp genome of M. ofeliae reported in this study will play an important role in understanding the evolution and diversification of the family. Furthermore, it will be an important genetic resource to the studies of conservation and restoration of M. ofeliae as an endangered Mexican magnolia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/ (reference number: MT522025) or available from the corresponding author.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Figlar RB, Nooteboom HP. 2004. Notes on Magnoliaceae IV. Blum. J Plant Tax Plant Geog. 49(1):87–100.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rivers M, Beech E, Murphy L, Oldfield S. 2016. The red list of Magnoliaceae. Revised and extended. Richmond (UK): Botanic Gardens Conservation International.
- Vázquez-García JA, Muñiz-Castro MÁ, Arroyo F, Pérez ÁJ, Serna M, Cuevas-Guzmán R, Domínguez-Yescas R, Castro-Arce E, Gurrola-Díaz CM. 2013. Novelties in neotropical Magnolia and an addendum proposal to the IUCN red list of Magnoliaceae. In: Salcedo-Pérez E, Hernández-Álvarez E, Vázquez-García JA, Escoto-García T, Díaz-Echavarría N, editors. Recursos forestales en el occidente de México, Serie Fronteras de Biodiversidad. Universidad de Guadalajara, Guadalajara. Vol. 4. p. 461–496.
- Vázquez-García JA, Neill DA, Asanza M, Pérez AJ, Arroyo F, Dahua-Machoa A, Merino-Santi RE. 2016. Magnolias de Ecuador: en riesgo de extinción. Puyo (Ecuador): Universidad Estatal Amazónica.
- Wang YB, Liu BB, Nie ZL, Chen HF, Chen FJ, Figlar RB, Wen J. 2020. Major clades and a revised classification of Magnolia and Magnoliaceae based on whole plastid genome sequences via genome skimming. J Syst Evol. 58(5):673–623.
