Abstract
We have determined the second mitochondrial genome of Epanerchodus koreanus Verhoeff, 1937 collected in limestone cave of Korea. The circular mitochondrial genome of E. koreanus is 15,581 bp long. It includes 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. Its gene order was different from the rest three Polydesmida mitochondrial genomes, resulted from relocation of tRNAs, rRNAs, and ND1. The base composition was AT-biased (75.1%). Phylogenetic trees displayed phylogenetic relationship, which is congruent to previous study, except Sphaerotheriidae sp. clustering with Helminthomorpha.
The genus Epanerchodus Attems, 1901 is comprising more than 70 species in the eastern Palearctic region (Golovatch Citation2014). Total eight millipedes belonging to the genus Epanerchodus Attems, 1901 have been reported in Korea (Mikhaljova and Lim Citation2006). Epanerchodus koreanus Verhoeff, 1937 (Polydesmidae: Polydesmida) is presented in both Korea and Asian part of Russia (Nguyen et al. Citation2016). In ecological aspects, E. koreanus is detritivores, and the most typical habitats of E. koreanus are leaf litter, the litter/soil, and around the organic matter at both entrance and twilight zone in a cave (Golovatch and Kime Citation2009; Golovatch Citation2015; Choi et al. Citation2016).
We completed the first mitochondrial genome of E. koreanus, collected in limestone cave, Ssang Cave, located in Mitan-myeon, Pyeongchang-gun, Gangwon-do, Republic of Korea (37°18′25.1ʺN, 128°31′58.6ʺE; its DNA were deposited at InfoBoss Cyber Herbarium (IN), IBS-00016). DNA was extracted using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences from Illumina NovaSeq6000 (Macrogen, Korea) were filtered by Trimmomatic v0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet v1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr/). Geneious R11 v11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate based on three mitochondrial genomes of Appalachioria falcifera (NC_021933; Brewer et al. Citation2013), Asiomorpha coarctata (KU721885; Dong et al. Citation2016), and Xystodesmus sp. (KU721886; Dong et al. Citation2016).
Epanerchodus koreanus mitochondrial genome (GenBank accession is MT898420) was 15,581 bp long. It contained 13 protein-coding genes (PCGs), 37 tRNAs, and two rRNAs and its AT ratio was 75.1%. Gene order of E. koreanus mitogenome was different from those of the three Polydesmida mitochondrial genomes, resulted from both tRNA relocations, commonly found in Myriapoda subphylum (Brewer et al. Citation2013), and rearrangement of 16S rRNA, ND4L, and 12S rRNA.
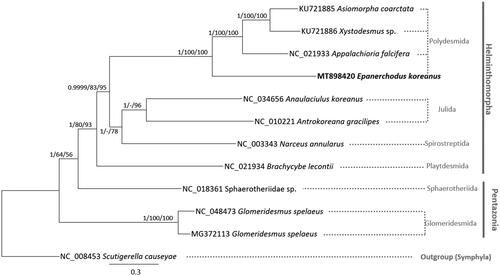
We inferred the phylogenetic relationship of eleven Diplopoda mitochondrial genomes including E. koreanus mitochondrial genome and one outgroup species, Scutigerella causeyae (NC_008453) based on concatenated alignments of 13 conserved PCGs by MAFFT v7.450 (Katoh and Standley Citation2013). Bootstrapped Bayesian inference, maximum-likelihood, and neighbor-joining phylogenetic trees were constructed with MrBayes v3.2.7a (Huelsenbeck and Ronquist Citation2001) and MEGA X (Kumar et al. Citation2018). Phylogenetic trees displayed that E. koreanus was clustered in Polydesmida clade with high supportive values (). In addition, they also presented phylogenetic relationship which was overall congruent to the previous studies (Sierwald and Bond Citation2007; Blanke and Wesener Citation2014), except Sphaerotheriidae sp. (NC_018361; Hu et al. Citation2020). Sphaerotheriidae sp. (NC_018361; Hu et al. Citation2020) belongs to infraclass Pentazonia, however, it clustered with infraclass Heminthomorpha in our phylogenetic tree (). We expected that out mitochondrial genome of E. koreanus which is a new species recently identified based on morphological characters in the eastern Palearctic region (Golovatch Citation2013, Citation2014) can be a fundamental genetic information for understanding species diversity and distribution of millipedes in the genus Epanerchodus.
Figure 1. Bayesian inference (1,000,000 generations), maximum-likelihood (10,000 bootstrap repeats), and neighbor-joining (10,000 bootstrap repeats) phylogenetic trees of eleven Diplopoda, and one Symphlya mitochondrial genomes as an out group: Epanerchodus koreanus (MT898420; This study), Asiomorpha coarctata (KU721885; Dong et al. Citation2016), Xystodesmus sp. (KU721886; Dong et al. Citation2016), Appalachioria falcifera (NC_021933; Brewer et al. Citation2013), Anaulaciulus koreanus (NC_034656; Woo HJ et al. Citation2017), Antrokoreana gracilipes (NC_010221; Woo H-J et al. Citation2007), Narceus annularus (NC_003343; Lavrov et al. Citation2002), Brachycybe lecontii (NC_021934; Brewer et al. Citation2013), Sphaerotheriidae sp. (NC_018361; Dong et al. Citation2012), Glomeridesmus spelaeus (NC_048473; unpublished), Glomeridesmus spelaeus (MG372113; Nunes et al. Citation2017), and one Symphlya species, Scutigerella causeyae (NC_008453; Podsiadlowski et al. Citation2007). Phylogenetic tree was drawn based on bayesian inference phylogenetic tree. The numbers above branches indicated posterior probability value of Bayesian inference, and bootstrap support values of maximum-likelihood and neighbor-joining phylogenetic trees, respectively. Order names were displayed as light gray color and infraclass names were written as gray color. In outgroup, class name was displayed as gray color.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MT898420 in NCBI GenBank.
Additional information
Funding
References
- Blanke A, Wesener T. 2014. Revival of forgotten characters and modern imaging techniques help to produce a robust phylogeny of the Diplopoda (Arthropoda, Myriapoda). Arthropod Struct Dev. 43(1):63–75.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Brewer MS, Swafford L, Spruill CL, Bond JE. 2013. Arthropod phylogenetics in light of three novel millipede (Myriapoda: Diplopoda) mitochondrial genomes with comments on the appropriateness of mitochondrial genome sequence data for inferring deep level relationships. PLOS One. 8(7):e68005
- Choi Y, Min H, An J, Paek W. 2016. A guide book of cave creatures. Daejeon (Korea): National Science Museum.
- Dong Y, Xu J-J, Hao S-J, Sun H-Y. 2012. The complete mitochondrial genome of the giant pill millipede, Sphaerotheriidae sp. (Myriapoda: Diplopoda: Sphaerotheriida)). Mitochondrial DNA. 23(5):333–335.
- Dong Y, Zhu L, Bai Y, Ou Y, Wang C. 2016. Complete mitochondrial genomes of two flat-backed millipedes by next-generation sequencing (Diplopoda, Polydesmida). ZooKeys. 637:1–20.
- Golovatch SI. 2013. Two new and one little-known species of the millipede genus Epanerchodus Attems, 1901 from southern China (Diplopoda, Polydesmida, Polydesmidae). Fragm Faun. 56(2):157–166.
- Golovatch SI. 2014. Review of the millipede genus Epanerchodus Attems, 1901 in continental China, with descriptions of new species (Diplopoda: Polydesmidae). Zootaxa. 3760(2):275–288.
- Golovatch SI. 2015. Cave Diplopoda of southern China with reference to millipede diversity in Southeast Asia. ZooKeys. 510:79–94.
- Golovatch SI, Kime RD. 2009. Millipede (Diplopoda) distributions: a review. Soil Organisms. 81(3):565–597.
- Hu C, Wang S, Huang B, Liu H, Xu L, Hu Z, Liu Y. 2020. The complete mitochondrial genome sequence of Scolopendra mutilans L. Koch, 1878 (Scolopendromorpha, Scolopendridae), with a comparative analysis of other centipede genomes. ZooKeys. 925(73):73–88.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lavrov DV, Boore JL, Brown WM. 2002. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss. Mol Biol Evol. 19(2):163–169.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Mikhaljova EV, Lim K-Y. 2006. The millipede genus Epanerchodus Attems, 1901 in the Korean Peninsula, with a description of a new species (Diplopoda, Polydesmida, Polydesmidae). Zootaxa. 1350(1):45–53.
- Nguyen AD, Jang KH, Hyun JS, Hwang UW. 2016. An Updated Checklist and Perspective Study of Millipedes (Arthropoda: Myriapoda: Diplopoda) in the Korean Peninsula. Anim System Evol Divers. 32(1):44–48..
- Nunes GL, Oliveira RRM, Pires ES, Vasconcelos S, Pietrobon T, Prous X, Oliveira G. 2017. Complete mitochondrial genome of Glomeridesmus spelaeus (Diplopoda), a troglobitic species from Carajás iron-ore caves (Pará, Brazil). BioRxiv. :228882.
- Podsiadlowski L, Kohlhagen H, Koch M. 2007. The complete mitochondrial genome of Scutigerella causeyae (Myriapoda: Symphyla) and the phylogenetic position of Symphyla. Mol Phylogenet Evol. 45(1):251–260.
- Sierwald P, Bond JE. 2007. Current status of the myriapod class Diplopoda (millipedes): taxonomic diversity and phylogeny. Annu Rev Entomol. 52:401–420.
- Woo H-J, Lee Y-S, Park S-J, Lim J-T, Jang K-H, Choi E-H, Choi Y-G, Hwang UW. 2007. Complete mitochondrial genome of a troglobite millipede Antrokoreana gracilipes (Diplopoda, Juliformia, Julida), and juliformian phylogeny. Mol Cells. 23(2), 574–583.
- Woo HJ, Nguyen AD, Jang KH, Choi EH, Ryu SH, Hwang UW. 2017. The complete mitochondrial genome of the Korean endemic millipede Anaulaciuluskoreanus (Verhoeff, 1937), with notes on the gene arrangement of millipede orders. Zootaxa. 4329(6):574–583.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
