Abstract
Alstroemeria, a member of the Alstroemeriaceae family, is a species from South America. The chloroplast genome of Alstroemeria spp. was completed by de novo assembly using a small amount of whole genome sequencing data. The chloroplast genome of Alstroemeria spp. was 155,672 bp in length consisting of 84,379 bp of large single copy, 17,815 bp of small single copy, and 26,739 bp of a pair of inverted repeat regions. A total of 157 genes were annotated including 103 protein-coding genes (PCGs), 46 tRNA genes, and eight rRNA genes. Maximum likelihood phylogenetic analysis with seven species belonging to the Alstroemeriaceae or Liliaceae family revealed that Alstroemeria spp. is grouped with the species in the Alstroemeriaceae family.
Alstroemeria belonging to the Alstroemeriaceae family is a popular cut flower plant because of the wide variety of flower colors and includes approximately 60 species (Aker and Healy Citation1990). The native species are found with many different shades of reds and yellow colors in numerous habitats and the wide color range in the interspecific hybrids arises from this diverse color base found in the species. The white and yellow types of color originated from one interspecific cross and the red and orange types of color originated from several different interspecific crosses (Healy and Wilkins Citation1986). Interspecific breeding is considered as the most important source of genetic variation in ornamental crops including Alstroemeria. However, albino progenies and variegated-color phenotypes caused by plastome-genome incompatibility appear during interspecific breeding in diverse plants, such as the genus, Pelargonium (Metzlaff et al. Citation1982), Trifolium (Pandey et al. Citation1987), Zantedeschia (Yao et al. Citation1994, Citation1995), etc. Therefore, the information of chloroplast genome of the hybrid ‘Hanhera’ (Alstroemeria spp.) obtained in this study will provide an insight of plastid inheritance, investigate more detailed evolutionary aspect, and facilitate breeding of colored Alstroemeria efficiently.
The Alstroemeria variety ‘Hanhera’ was provided by Prof. Tae-Ho Han, Department of Horticulture, Chonnam National University, South Korea (35.2° N, 126.9° E). It is an Alstroemeria hybrid (Alstroemeria spp.) derived from a cross between other Alstroemeria spp. hybrids ‘Leon’ and ‘Everest’. Their genetic backgrounds are complex because they are originally derived from interspecific crosses between different species belonging to the Alstroemeria genus. The variety ‘Handera’ has white-colored flowers with very faint pink and yellow petal. Its flower size is bigger and plant height is taller than other Alstroemeria varieties. The completion of the chloroplast genome sequence was performed at Phyzen bioinformatics pipeline (Kim et al. Citation2015). According to the PE standard protocol (Illumina, San Diego, CA), an Illumina paired-end (PE) genomic library was constructed with a total genomic DNA and they were sequenced using an Illumina HiSeq2000 platform at Macrogen (http://www.macrogen.com/kor/). The chloroplast genome assembly was performed with approximately 1.9 Gbp of trimmed high-quality reads obtained by a CLC assembly cell package version 4.2.1 (CLC Inc., Aarhus, Denmark). The principal contigs representing the chloroplast genome were retrieved using Nucmer (Kurtz et al. Citation2004) with the chloroplast genome sequence of Alstroemeria aurea (KC968976, Do et al. Citation2013; Kim and Kim Citation2013) as reference sequence. The representative chloroplast contigs were arranged in order based on BLASTZ analysis (Schwartz et al. Citation2003) and connected to a single draft sequence by joining overlapping terminal sequences and manual editing through a comparison with the reference chloroplast genome sequence of Alstroemeria aurea (KC968976) as described previously (Cho et al. Citation2015; Cho and Park Citation2016). GeSeq program (Tillich et al. Citation2017) and manual curation based on the results of BLAST searches were used for gene annotation.
The complete chloroplast genome of the Alstroemeria hybrid variety ‘Hanhera’ (GenBank accession no. MK327552) was 155,672 bp in length including 26,739 bp inverted repeats (IRa and IRb) regions separated by small single copy (SSC) region of 17,815 bp and large single copy (LSC) region of 84,379 bp. When the chloroplast genome sequence was compared with those of other Alstroemeriaceae species, such as A. aurea (KC968976, Do et al. Citation2013; Kim and Kim Citation2013) and Bomarea edulis (KM233641, Kim et al. Citation2016), it was slightly longer and the sequence conservation between them was expectedly very high with a sequence identity of 99 and 96%, respectively. The structure was quadripartite that is typical in most plastids, and the structure and gene features were typically identical to those of higher plants. A total of 157 genes with an average size of 588.2 bp were annotated including 103 protein-coding genes (PCGs) with an average size of 780.9 bp, 46 tRNA genes, and eight rRNA genes with an average size of 220.7 bp. An overall GC content was 38.05%.
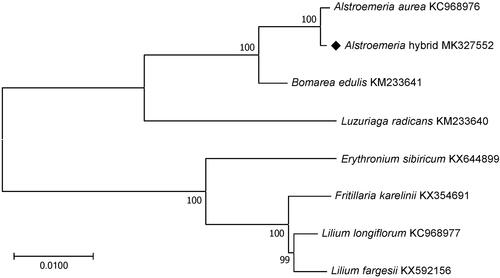
The results from the phylogenetic analysis performed using chloroplast coding sequences of the Alstroemeria hybrid ‘Hanhera’ and three and four published species in the Alstroemeriaceae and Liliaceae family, respectively, by a maximum likelihood method in MEGA version 6.0 (Tamura et al. Citation2013) revealed that the Alstroemeria hybrid ‘Hanhera’ was expectedly grouped with A. aurea followed by Bomarea edulis and Luzuriaga radicans which all belonged to the Alstroemeriaceae family ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MK327552.
Additional information
Funding
References
- Aker S, Healy W. 1990. The phytogeography of the genus Alstroemeria. Herbertia. 46:76–87.
- Cho KS, Park TH. 2016. Complete chloroplast genome sequence of Solanum nigrum and development of markers for the discrimination of S. nigrum. Hortic Environ Biotechnol. 57(1):69–78.
- Cho KS, Yun BK, Yoon YH, Hong SY, Mekapogu M, Kim KH, Yang TJ. 2015. Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PLOS One. 10(5):e0125332.
- Do HDK, Kim JS, Kim JH. 2013. Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae). Gene. 530(2):229–235.
- Healy WE, Wilkins HF. 1986. Alstroemeria culture. Herbertia. 42:16–20.
- Kim JS, Kim HT, Yoon CY, Kim JH. 2016. The complete plastid genome sequence of Bomarea edulis (Alstroemeriaceae: Liliales). Mitochondrial DNA Part A. 27:1869–1870.
- Kim JS, Kim JH. 2013. Comparative genome analysis and phylogenetic relationship of order Liliales insight from the complete plastid genome sequences of two Lilies (Lilium longiflorum and Alstroemeria aurea). PLoS One. 8(6):e68180.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.
- Metzlaf M, Pohlheim F, Börner T, Hagemann R. 1982. Hybrid variegation in the genus Pelargonium. Curr Genet. 5(3):245–249.
- Pandey KK, Grant JE, Williams EG. 1987. Interspecific hybridaisation between Trifolium repens and T. uniflorum. Aust J Bot. 35(2):171–182.
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13(1):103–107.
- Tamura K, STecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yao JL, Cohen D, Rowland RE. 1994. Plastid DNA inheritance and plastome-genome incompatibility in interspecific hybrids of Zantedeschia (Araceae). Theor Appl Genet. 88(2):255–260.
- Yao JL, Cohen D, Rowland RE. 1995. Interspecific albino and variegated hybrids in the genus Zantedeschia. Plant Sci. 109(2):199–206.