Abstract
In this study, we determined the mitochondrial genome of a stomatopod, Chorisquilla orientalis, collected from Korean waters. The complete mitochondrial genome comprised 15,880 bp, encoding 13 proteins, 22 transfer RNAs, 2 ribosomal RNAs genes, and a non-coding A + T rich region. The overall base composition in the heavy strand was A: 35.5%, G: 12.4%, C: 18.3%, and T: 33.7%, with a G + C content of 30.8%. Phylogenetic analysis showed that C. orientalis belonged to the families Gonodactylidae and Takuidae in the same clade, and to the superfamily Gonodactyloidea within Stomatopoda. This is the first record of the complete mitochondrial genome sequence of the family Protosquillidae.
Mantis shrimps or stomatopods have large and powerful raptorial appendages that can be used for ‘smashing’ or ‘spearing’ (Caldwell and Dingle Citation1976). More than 480 stomatopod species have been described worldwide (Ahyong Citation2001, Citation2004), they play important roles in numerous marine ecosystems, due to their enormous biomass and nutritional position as both prey and predator (Geary et al. Citation1991; Ahyong et al. Citation2013). Of them, all protosquillids have a smashing-type of raptorial claw and include more than 35 species in 6 genera have been reported (Ahyong Citation2010). In this study, we provide the first complete mitochondrial genome sequence of a Protosquillid species, Chorisquilla orientalis Hwang, Ahyong, & Kim.
The specimen was collected by scuba diving from the subtidal zone of Dokdo Island, South Korea (geographic location: 37°14′34.9′′N, 131°52′08.6′′E) on 16 July 2018, and was preserved in 95% ethyl alcohol until mitogenome analysis. The voucher specimen was deposited at the Research Institute of EcoScience, Ewha Womans University (EWNHMAR769). Total DNA was extracted from leg muscle tissue using DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) and the DNA library was prepared using TruseqNano DNA Prep Kit (Illumina, San Diego, CA). The mitochondrial DNA (mtDNA) was sequenced using Illumina Novaseq 6000 system. MITObim (Hahn et al. Citation2013) was used for the assembly of the complete mitochondrial genome, which was then annotated using MITOS (Bernt et al. Citation2013).
The length of the mitogenome of C. orientalis is 15,880 bp encoding 13 proteins, 22 transfer RNAs, 2 ribosomal RNAs, and a non-coding A + T-rich control region. For the 13 protein-coding genes (PCGs), the most common shared start codon was ATG in COX2, COX3, ATP6, NAD3, NAD4, NAD4L, and CYTB, followed by ATT in NAD2 and NAD6. The start codon for COX1 was ACG, which has also been reported in the mtDNA of malacostraca (Cook Citation2005; Liu and Cui Citation2010), while NA1 and NAD5 started with ATA, and ATP8 with an ATC. The stop codon in all PCGs was TAA, except for COX2 and NAD6 that ended with the incomplete stop codons AAT and CCT, respectively. Incomplete stop codons have been identified in several PCGs of all the stomatopod mitochondrial genomes published to date, and this has been attributed to excessive polyadenylation (Ojala et al. Citation1980, Citation1981). The overall base composition in the heavy strand was A: 35.5%, G: 12.4%, C: 18.3%, and T: 33.7%, with a G + C content of 30.8%. The LrRNA and SrRNA genes in C. orientalis had 1353 bp and 838 bp, respectively. The size of the 22 transfer RNAs encoded ranged from 64 to 72 nucleotides. The putative control region was of 947 bp and was located after the transfer RNA-Val and the SrRNA.
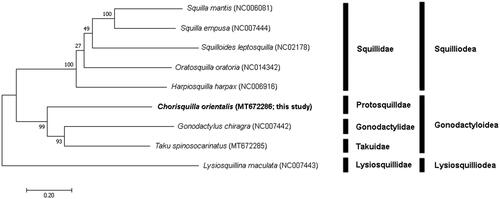
The phylogenetic tree was constructed based on sequences of 13 PCGs identified by the maximum likelihood (ML) method using MEGA X (Kumar et al. Citation2018). The GTR + G + I model was identified as the best-fit model for the data, using ModelFinder (Kalyaanamoorthy et al. Citation2017) with 1,000 bootstrap replicates.
To confirm the phylogenetic position of C. orientalis, four squilloid species and two gonodactyloid were compared with our C. orientalis (MT672286), using a lysiosquilloid species as an outgroup. The analysis showed that C. orientalis was grouped with Gonodactylus chiragra of Gonodactylidae and Taku spinosocarinatus of Takuidae, in one clade with high bootstrap value, and it belonged to the superfamily Gonodactyloidea within Stomatopoda ().
Figure 1. Phylogenetic tree of complete mitochondrial genomes from nine stomatopods (Oratosquilla oratoria (NC014342), Gonodactylus chiragra (NC007442), Harpiosquilla harpax (NC006916), Squilla empusa (NC007444), Squilla mantis (NC006081) Lysiosquillina maculata (NC007443), Taku spinosocarinatus (MT672285), and Chorisquilla orientalis (MT672286)) constructed using maximum likelihood (ML) method.
This is the first report of the complete mitogenome sequence of the family Protosquillidae. The results of this study provide useful information for further phylogenetic and evolutionary studies on stomatopod members in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MT672286. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA663208, SRR12649330, and SAMN16122871, respectively. The data that support the findings of this study are also openly available in Mendeley Data at http://dx.doi.org/10.17632/khwyfjmt7g.1
Additional information
Funding
References
- Ahyong ST. 2001. Revision of the Australian stomatopod Crustacea. Rec Austral Museum Suppl. 26:1–326.
- Ahyong ST. 2004. Stomatopoda – mantis shrimps. In: Poore GCB, editor. Marine decapod Crustacea of Southern Australia: a guide to identification. Victoria, Australia: CSIRO Publishing, Colling-Wood; p. 517–549.
- Ahyong ST. 2010. A new genus and two new species of mantis shrimp from the Western Pacific (Stomatopoda: Gonodactyloidea: Protosquillidae). J Crustacean Biol. 30 (1):141–145.
- Ahyong ST, Charbonnier S, Garassino A. 2013. Squilla taulinanus n. sp. (Crustacea, Stomatopoda, Squillidae) from the Burdigalian (Miocene) of Taulignan, southeastern France. Bolet Soc Geol Mexica. 65(2):213–217.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Caldwell RL, Dingle H. 1976. Stomatopods. Sci Am. 234(1):80–89.
- Cook CE. 2005. The complete mitochondrial genome of the stomatopod crustacean Squilla mantis. BMC Genomics. 6:105.
- Geary DH, Allmon WD, Reaka-Kudla ML. 1991. Stomatopod predation on fossil gastropods from the Plio-Pleistocene of Florida. J Paleontol. 65(03):355–360.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu Y, Cui Z. 2010. The complete mitochondrial genome of the mantid shrimp Oratosquilla oratoria (Crustacea: Malacostraca: Stomatopoda): novel non-coding regions features and phylogenetic implications of the Stomatopoda. Comp Biochem Physiol Part D Genom Proteom. 5(3):190–198.
- Ojala D, Merkel C, Gelfand R, Attardi G. 1980. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 22(2 Pt 2):393–403.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
