Abstract
In this study, we report the mitochondrial genome of the black-tailed antechinus (Antechinus arktos), a recently-discovered, endangered carnivorous marsupial inhabiting a caldera that straddles the border of Australia’s mid-east coast. The circular A. arktos genome is 17,334 bp in length and has an AT content of 63.3%. Its gene content and arrangement are consistent with reported marsupial mitogenome assemblies.
Here, we present the complete mitochondrial genome of the black-tailed dusky antechinus (Antechinus arktos). Described in 2014, A. arktos is a rare species, apparently limited to the highest, wettest reaches of the Tweed Volcano Caldera in mid-eastern Australia (Baker et al. Citation2014). In 2018, the species was federally listed as Endangered, and it was recently recognized as one of the top 20 Australian mammals most likely to go extinct in the next two decades (Geyle et al. Citation2018).
Genomic DNA was extracted from ear tissue (voucher specimen AA100) collected from Lamington National Park, Queensland, Australia (28.26S, 153.17E). Paired-end 350 bp-insert DNA libraries were sequenced by BGI (Hong Kong), using the BGISEQ-500 instrument, to generate ∼30× genome coverage of 100 bp paired-end reads. Raw data was processed as described in a recent ‘mitocommunication’ on Murexia melanurus (Tian et al. Citation2019).
The mitogenome was assembled as follows: 48 M reads were assembled using NOVOPlasty version 2.7.2 (Dierckxsens et al. Citation2017), with the ND1 coding sequence from a partial Antechinus flavipes mitogenome (GenBank accession no. KJ868098) (Mitchell et al. Citation2014) as a seed sequence and the parameters ‘Type = mito K-mer = 23 Genome range = 15000–19000 Variance detection = no.’ A. arktos reads were present at high coverage (∼1000× across the mitogenome) and assembled to give a single contig. NOVOPlasty indicated that two adjacent nucleotides (out of 17,334) could not be accurately called (likely due to their presence within a repeat region; indicated by ‘*’ in the output FASTA). Geneious Prime version 2019.1.3 (Biomatters, Auckland, New Zealand) was used to align a further 47M reads against the assembled contig and generate a consensus genome sequence – settings ‘Majority Threshold’ and a minimum calling coverage of 100 – resolving the ambiguous nucleotides in the assembly. The genome was annotated using GenBank features of the Northern quoll (Dasyurus hallucatus; accession no. NC_007630). We used MARS (Ayad and Pissis Citation2017) to rotate the sequences to the same origin.
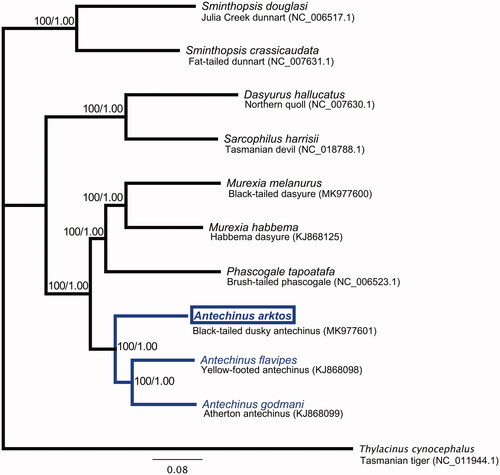
The final circular A. arktos mitochondrial genome (GenBank: MK977601) is 17,334 bp in length and has a base composition of 32.8% A, 30.5% T, 12.9% G, and 23.8% C. As in other mammals, the genome has 13 protein-coding genes (PCGs) and two ribosomal (rRNA genes). The genome shares unique features with all marsupial mitogenomes reported to date. These include a ‘ACWNY’ tRNA gene re-arrangement (Paabo et al. Citation1991); 21 transfer RNA (tRNA), due to a tRNALys pseudogene (no lysine anticodon) (Janke et al. Citation1994; Dorner et al. Citation2001); and lack of an aspartic acid anticodon, likely post-transcriptionally rescued by RNA-editing (Janke and Paabo Citation1993). In agreement with previous reports (Krajewski et al. Citation2007; Kumar et al. Citation2017; Mutton et al. Citation2019), phylogenetic analysis revealed that Antechinus is closely related to the genera Phascogale of Australia and Murexia of New Guinea, and A. arktos forms a monophyletic group along with previously reported Antechinus species (). Maximum-likelihood (ML; assessed using IQ-TREE (Nguyen et al. Citation2015)) and Bayesian Interference (BI; assessed using MrBayes version 3.2.7 (Ronquist and Huelsenbeck Citation2003)), with the mtMAM+I+G4 model chosen using to the Bayesian Information Criterion) resulted in an identical tree topology.
Figure 1. Phylogenetic tree of black-tailed dusky antechinus (Antechinus arktos; indicated by a box), nine other species in the marsupial family Dasyuridae, and the outgroup species Thylacinus cynocephalus. Phylogenetic reconstruction was performed with coding sequences of the 13 PCGs. The number at each node are ML/BI bootstrap support values.
Geolocation information
Geospatial coordinates for the black-tailed dusky antechinus (A. arktos) ear tissue collection: 28.26S, 153.17E.
Acknowledgments
We thank Drs. Emma Gray (Queensland University of Technology) and Ian Gynther (Queensland Department of Environment and Science) for providing the A. arktos sample.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study is available at NCBI (National Center for Biotechnology Information) https://www.ncbi.nlm.nih.gov GenBank (accession no. MK977601).
Additional information
Funding
References
- Ayad LA, Pissis SP. 2017. MARS: improving multiple circular sequence alignment using refined sequences. BMC Genomics. 18(1):86.
- Baker AM, Mutton TY, Hines HB, Van Dyck S. 2014. The black-tailed antechinus, Antechinus arktos sp. nov.: a new species of carnivorous marsupial from montane regions of the Tweed Volcano caldera, eastern Australia. Zootaxa. 3765(2):101–133.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Dorner M, Altmann M, Paabo S, Morl M. 2001. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol Biol Cell. 12(9):2688–2698.
- Geyle HM, Woinarski JC, Baker GB, Dickman CR, Dutson G, Fisher DO, Ford H, Holdsworth M, Jones ME, Kutt A. 2018. Quantifying extinction risk and forecasting the number of impending Australian bird and mammal extinctions. Pac Conserv Biol. 24(2):157–167.
- Janke A, Feldmaier-Fuchs G, Thomas WK, von Haeseler A, Pääbo S. 1994. The marsupial mitochondrial genome and the evolution of placental mammals. Genetics. 137(1):243–256.
- Janke A, Paabo S. 1993. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 21(7):1523–1525.
- Krajewski C, Torunsky R, Sipiorski JT, Westerman M. 2007. Phylogenetic relationships of the dasyurid marsupial genus Murexia. J Mammal. 88(3):696–705.
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. Time tree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819.
- Mitchell KJ, Pratt RC, Watson LN, Gibb GC, Llamas B, Kasper M, Edson J, Hopwood B, Male D, Armstrong KN, et al. 2014. Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Mol Biol Evol. 31(9):2322–2330.
- Mutton TY, Phillips MJ, Fuller SJ, Bryant LM, Baker AM. 2019. Systematics, biogeography and ancestral state of the Australian marsupial genus Antechinus (Dasyuromorphia: Dasyuridae). Zool J Linn Soc. 186(2):553–568.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Paabo S, Thomas WK, Whitfield KM, Kumazawa Y, Wilson AC. 1991. Rearrangements of mitochondrial transfer RNA genes in marsupials. J Mol Evol. 33(5):426–430.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Tian R, Geng Y, Thomas PB, Jeffery PL, Mutton TY, Chopin LK, Baker AM, Seim I. 2019. The mitochondrial genome of the black-tailed dasyure (Murexia melanurus). Mitochondrial DNA Part B. 4(2):3598–3600.
