Abstract
The mitochondrial genome of Sargassum patens was completely sequenced. This mitogenome is a circular molecule of 34,844 bp in length and had an overall GC content of 36.20%. The mitogenome contains 37 protein-coding, three rRNA, 25 tRNA genes and two conserved open reading frames (ORFs). The Maximum-likelihood (ML) tree showed that S. patens has a closer relationship with S. aquifolium.
Sargassum patens C. Agardh (Sargassaceae, Phaeophyceae) is a perennial macroalgae, known widely distributed on rocky subtidal shores from the Japanese and Chinese seas (Huang et al. Citation2013). The detached thalli of S. patens had a floating period of between 1 and 9 weeks (Yatsuya Citation2008), which could provide food and habitat for fish larva and invertebrates in pelagic ecosystem (Yamasaki et al. Citation2014). We described the characterization of complete mitogenome of S. patens which were expected to facilitate future studies on taxonomic resolution, population genetic structure and phylogenetic relationships.
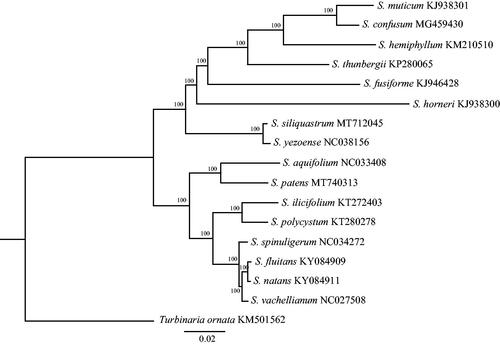
The specimen was collected from Bailong Island, Guangxi Province, China (21°30′20″N; 108°13′46″E) on 20 January 2020, and stored at the Marine Biological Museum, Chinese Academy of Sciences with an accession number MBM286786. Paired-end libraries with insert sizes of ∼400 bp were prepared and used for Illumina HiSeq 2000 sequencing. The raw paired end reads were trimmed and quality controlled by Trimmomatic (Bolger et al. Citation2014). ABySS was used to perform genome assembly (Simpson et al. Citation2009), and GapCloser software (https://anaconda.org/bioconda/soapdenovo2-gapcloser) was applied to fill up the remaining local inner gaps and corrected the single base polymorphism. The transfer RNA (tRNA) genes were predicted by tRNAscan-SE (Lowe and Chan Citation2016) and the ribosomal RNA (rRNA) genes and protein coding genes were determined based on homology alignments with Sargassum aquifolium (NC033408). To explore the phylogenetic position of S. patens, mitochondrial genome sequences of 16 species (downloaded from the GenBank database) were used to construct phylogenetic tree by the Maximum-likelihood (ML) method, with Turbinaria ornata (GenBank accession number KM501562) served as the out-group.
The complete mitogenome (MT740313) comprises a circular DNA molecule measuring 34,844 bp in length with 36.20% overall GC content. The mitogenome contains 65 genes, including 37 protein-coding, three rRNA, 25 tRNA genes and two conserved open reading frames (ORFs). All protein-coding genes use the start codon ATG. Of the 37 protein-coding genes, 27 ended with the TAA stop codon, five with TAG and five with TGA. The lengths of three rRNA genes are 1939 bp (rnl rRNA), 874 bp (rns rRNA), and 126 bp (rrn5 rRNA). All genes show the typical gene arrangement conforming to the Sargassaceae family consensus (Liu et al. Citation2019). Phylogeographic tree showed that S. patens clustered together with S. aquifolium (). The complete mitogenome sequence provided herein would help understand molecular evolution of the related species of Sargassaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in repository under Accession: MT740313. https://www.ncbi.nlm.nih.gov/nuccore/1898549977. Raw sequence data were submitted to the NCBI SRA (BioProject: PRJNA668679).
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics. 30(15):2114–2120.
- Huang BX, Ding LP, Tan HQ, Sun GD. 2013. Species diversity and distribution of genus Sargassum in China seas. Oceanologia et Limnologia. 44:69–76.
- Liu F, Zhang Y, Bi Y, Chen W, Moejes FW. 2019. Understanding the evolution of mitochondrial genomes in Phaeophyceae inferred from mitogenomes of Ishige okamurae (Ishigeales) and Dictyopteris divaricata (Dictyotales). J Mol Evol. 87(1):16–26.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6):1117–1123.
- Yamasaki M, Aono M, Ogawa N, Tanaka K, Imoto Z, Nakamura Y. 2014. Drifting algae and fish: implications of tropical Sargassum invasion due to ocean warming in western Japan. Estuar. Coast Shelf S. 147:32–41.
- Yatsuya K. 2008. Floating period of Sargassacean thalli estimated by the change in density. J Appl Phycol. 20(5):797–800.