Abstract
In this study, the complete 16,184 bp mitochondrial genome of Cyclograpsus intermedius was determined from a specimen collected in South Korea. It consists of 13 protein-coding, 22 tRNA, 2 rRNA genes, and a non-coding A + T rich region. The base composition of the heavy strand in the mitochondrial genome was 34.7% A, 10.7% G, 18.7% C, and 35.9% T, resulting in a G + C content of 29.4%. A maximum-likelihood phylogenetic tree based on the 13 mitochondrial protein-coding genes showed that C. intermedius clustered together with the Varunidae. These molecular data will be useful for studying the evolutionary relationships among crab species.
Grapsoidea is a superfamily of crabs comprising species that adapt to terrestrial, semi-terrestrial, or freshwater environments and plays an important role in the coastal ecosystem (Lee Citation1998). The species Cyclograpsus intermedius inhabits both temperate and tropical regions, including Korea, Japan, Taiwan, and the Indian Ocean (Hangai et al. Citation2009; Tan et al. Citation2016). Despite the discovery of morphologically similar species in the genus Cyclograpsus, genetic and taxonomic features of the genus remain uncharacterized (Griffin Citation1968; Hangai et al. Citation2009). We sequenced the mitochondrial genome of a Korean specimen of C. intermedius to construct the taxonomy and phylogeny of grapsid crabs; thus, providing more molecular data relating to this superfamily of crabs (GenBank accession number: MT621398).
A specimen of C. intermedius was collected from the rocky intertidal zone of Dokdo, South Korea on 20 September 2019 (geographic location: 37°14′29.6′′ N, 131°52′10.3′′ E). The specimen was preserved in 80% ethanol and stored at the Ewha Womans University Natural History Museum in Korea (accession number: EWNHMMAR767). Total DNA was extracted from the muscle of the dissected walking legs of the specimen using DNeasy Blood & Tissue (Qiagen, Valencia, CA). The mtDNA was sequenced using the Novaseq 6000 System (Illumina, San Diego, CA). The MITObim method (Hahn et al. Citation2013) and MITOS (Bernt et al. Citation2013) were used for the assembly and annotation of the complete mitochondrial genome, respectively.
The mitogenome of C. intermedius was 16,184 bp in length, which is a typical length of Decapoda mitogenomes. It included 13 protein-coding, 22 tRNA, 2 rRNA genes, and a non-coding A + T-rich control region. For the 13 protein-coding genes, the most common shared start codon was ATG (in COX1, COX2, COX3, ATP8, ND4L, and ND4), followed by ATT (ATP6, ND5, and ND6). The most common termination codon was TAA (COX1, COX3, ND1, ND3, ND4, ND4L, ND6, ATP6, and ATP8), followed by the incomplete termination codon T– (ND2, ND5, CYTB, and COX2). The overall mitochondrial base composition of this genome was A: 34.7%, T: 35.9%, G: 10.7%, and C: 18.7%, with a G + C content of 29.4%.
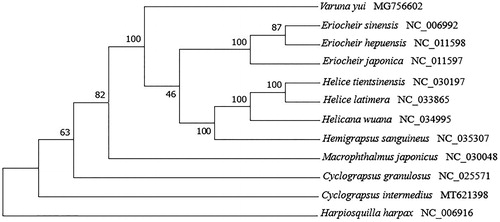
To determine the phylogenetic relationship of C. intermedius, a multiple sequence alignment was prepared by concatenating sequences of the 13 mitochondrial protein-coding genes from 11 crab species and an outgroup (Harpiosquilla harpax) in the NCBI GenBank database. A maximum-likelihood phylogenetic tree was constructed using MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replicates (). Cyclograpsus intermedius was clustered with other Varunidae species, suggesting that C. intermedius is also a member of Grapsoidea. In conclusion, the complete mitogenome of C. intermedius provides fundamental phylogenetic information of the genus Cyclograpsus.
Disclosure statement
No potential competing interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Mendeley Data at http://dx.doi.org/10.17632/jn3wh7bc74.1
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Griffin DJG. 1968. A new species of Cyclograpsus (Decapoda, Grapsidae) and notes on five others from the Pacific Ocean. Crustaceana. 15(3):235–248.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Hangai R, Kitaura J, Wada K, Fukui Y. 2009. A new species of Cyclograpsus (Brachyura: Varunidae) from Japan, co-occurring with C. intermedius Ortmann, 1894. Crust Res. 38(0):21–27.
- Lee SY. 1998. Ecological role of grapsid crabs in mangrove ecosystems: a review. Mar Freshwater Res. 49(4):335–343.
- Tan MH, Gan HM, Lee YP, Austin CM. 2016. The complete mitogenome of purple mottled shore crab Cyclograpsus granulosus H. Milne-Edwards, 1853 (Crustacea: Decapoda: Grapsoidea). Mitochondrial DNA Part A. 27(6):3981–3982.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.