Abstract
We de novo assembled the complete mitochondrial genome of the green peach aphid, Myzus persicae, using its genomic DNA isolated from the bell pepper in Korea. The circular mitogenome of M. persicae is 16,936 bp long and contains the standard 37 genes: 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes, as well as a single control region of 798 bp. Given the high AT ratio (84.1%) of the M. persicae mitogenome, we found, through the comparison of the Chinese M. persicae mitogenomes, that approximately 1.6% of the mitogenome is polymorphic, including 30 single nucleotide polymorphisms (SNPs), 12 insertions and deletions (INDELs), and large sequence variations in the control region. To resolve the phylogenetic position of M. persicae, we analyzed all mitochondrial protein-coding genes from 38 species within the Aphidoidea superfamily, with Adelges laricis as an outgroup. Our M. persicae sample was significantly grouped with three existing M. persicae samples, and the species belonging to the family Aphididae formed a monophyletic clade.
Myzus persicae (Sulzer, 1776) is a notorious pest of many agricultural crops worldwide and acts as a vector for the transport of plant viruses (Bass et al. Citation2014; Katis et al. Citation2017). Its development appears to be rapid, often taking 10–12 days for one generation, with at least more than 20 generations per year in mild climates. Eggs are initially yellow or green, but soon turn black. Similarly, initially greenish nymphs rapidly turn yellowish. The length of adult M. persicae body is 1.8–2.1 mm. Depending on the food conditions, it can exist in the viviparous summer stage that feeds widely or oviparous winter stage that has a restricted diet or experiences nutritional imbalance (Capinera Citation2000). Myzus persicae feeds on various host plants and intriguingly tends to be present at high densities on young plant tissues, causing leaf atrophy, slow growth, and reduced yield (Petitt and Smilowitz Citation1982). This species has been found to be resistant to various insecticides (Bass et al. Citation2014; Voudouris et al. Citation2017).
To investigate intra-specific variations and phylogenetic relationship of M. persicae, we extracted, amplified, and sequenced DNA from M. persicae isolated from bell pepper (Capsicum annuum) collected from Suwon, Gyeonggi-do, Republic of Korea; 35°50′26.8ʺN, 127°02′42.9ʺE; InfoBoss Cyber Herbarium (IN); INH-00025 using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). Sequencing library was constructed using Illumina TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, CA) following manufacturer’s recommendations with around 350-bp DNA fragments. 5.86 Gbp raw sequence reads were obtained from Illumina NovaSeq6000 (Macrogen Inc., South Korea) and preprocessed by Trimmomatic v0.33 (Bolger et al. Citation2014), and the remaining clean reads were used to de novo assemble the mitogenome using Velvet v1.2.10 (Zerbino and Birney Citation2008). To close gaps in the draft mitogenome, the environment of the Genome Information System (GeIS; http://geis.infoboss.co.kr; Park et al., in preparation) including SOAPGapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17, and SAMtools v1.9 (Li et al. Citation2009; Li Citation2013), was applied. Finally, the 16,936-bp complete mitogenome of M. persicae (GenBank accession MT900593) was obtained.
Using the Geneious R11 v11.1.5 software (Biomatters Ltd, Auckland, New Zealand) with the existing M. persicae mitogenome (NC_029727; Yang et al. Citation2017) as a reference, our newly assembled mitogenome was annotated with 13 protein-coding genes (PCGs), 2 rRNAs, and 22 tRNAs, and had a high AT ratio (84.1%). We investigated intraspecific mitogenomic variation based on the existing mitogenome isolated from Chinese M. persicae (Yang et al. Citation2017) and identified 30 single nucleotide polymorphisms (SNPs), 12 insertions and deletions (INDELs), and 3 large INDELs in the control region (with sizes 438, 220, and 233 bp). These intragenic variations are larger than those of Aphis gossypii (Park, Xi, et al. 2019; Bae et al. 2020), Nilaparvata lugens (Choi et al. Citation2019; Park, Kwon, et al. Citation2019; Choi et al. Citation2020), Laodelphax striatellus (Park, Jung, et al. Citation2019; Seo, Jung, et al. Citation2019), and Spodoptera frugiperda (Seo, Lee, et al. Citation2019), but are smaller than those of Chilo suppresallis (Park, Xi, et al. Citation2019).
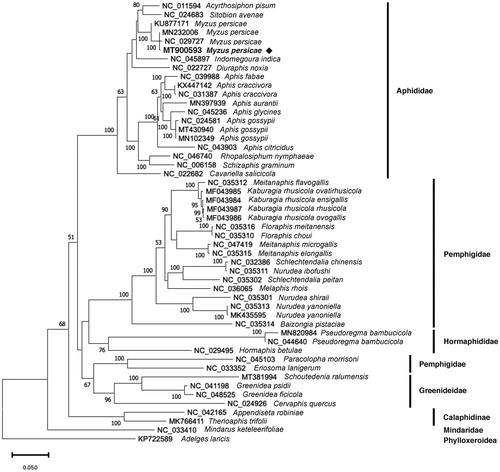
To resolve the phylogenetic position of M. persicae, we analyzed 13 PCGs from 38 species within the Aphidoidea superfamily with Adelges laricis as an outgroup. Multiple sequence alignment of each gene in all the samples was obtained using MAFFT v7.453 (Katoh and Standley Citation2013). These alignments were then concatenated using phyutility v2.7.1 (Smith and Dunn Citation2008). A maximum-likelihood phylogenetic tree was generated with IQ-TREE v1.6.12 (Nguyen et al. Citation2015) using a mtMet+F+R4 substitution model with 1,000 bootstrap replicates. We found that our Korean M. persicae mitogenome was clearly clustered with the existing M. persicae mitogenomes in a monophyletic manner, and it was closer to the Chinese M. persicae mitogenomes than the Brazilian M. persicae mitogenome (). In addition, the Aphididae family including M. persicae represented a clear monophyletic relationship, whereas the Pemphigidae family are paraphyletic (). In conclusion, our M. persicae mitogenome will provide a useful genetic resource and help to understand the phylogenetic relationship of the Aphidoidea clade.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MT900593 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA667955, SAMN16392923, and SRR12791240, respectively.
Additional information
Funding
References
- Bae Y, Park J, Lee W. 2020. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) isolated from Plantago asiatica in Korea. Mitochondrial DNA Part B. 5(3):2896–2898.
- Bass C, Puinean AM, Zimmer CT, Denholm I, Field LM, Foster SP, Gutbrod O, Nauen R, Slater R, Williamson MS. 2014. The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem Mol Biol. 51:41–51.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Capinera JL. 2000. melon aphid or cotton aphid – Aphis gossypii Glover. [accessed Jul 27, 2020]. http://entnemdept.ufl.edu/creatures/veg/aphid/melon_aphid.htm.
- Choi NJ, Lee B-C, Park J, Park J. 2019. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in China (Hemiptera: Delphacidae): investigation of intraspecies variations between countries. Mitochondrial DNA Part B. 4(1):1677–1678.
- Choi NJ, Lee B-C, Park J, Park J. 2020. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in Guangxi province, China (Hemiptera: Delphacidae): identification of the origin of N. lugens migrated to Korea. Mitochondrial DNA Part B. 5(2):1960–1961.
- Katis NI, Tsitsipis JA, Stevens M, Powel G. 2017. Transmission of plant viruses. Aphids as crop pests. 2nd ed. 714.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Park J, Jung JK, Ho Koh Y, Park J, Seo BY. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a mid-Western part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2229–2230.
- Park J, Kwon W, Park J, Kim H-J, Lee B-C, Kim Y, Choi NJ. 2019. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in Korea (Hemiptera: Delphacidae). Mitochondrial DNA Part B. 4(1):1674–1676.
- Park J, Xi H, Kim Y, Park J, Lee W. 2019. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) collected in Korean peninsula. Mitochondrial DNA Part B. 4(2):3007–3009.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863) (Lepidoptera: Crambidae). Mitochondrial DNA Part B. 4(1):850–851.
- Petitt FL, Smilowitz Z. 1982. Green peach aphid feeding damage to potato in various plant growth stages. J Econ Entomol. 75(3):431–435.
- Seo BY, Jung JK, Ho Koh Y, Park J. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a southern part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2242–2243.
- Seo BY, Lee G-S, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondrial DNA Part B. 4(2):3918–3920.
- Smith SA, Dunn CW. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716.
- Voudouris CC, Williamson MS, Skouras PJ, Kati AN, Sahinoglou AJ, Margaritopoulos JT. 2017. Evolution of imidacloprid resistance in Myzus persicae in Greece and susceptibility data for spirotetramat. Pest Manag Sci. 73(9):1804–1812.
- Yang H, Wang J, Yu H, Yang M. 2017. Sequence and phylogenetic analysis of the complete mitogenome of Myzus persicae (Hemiptera: Aphididae). Acta Entomologica Sinica. 60(1):84–94.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(Suppl 14):S2.