Abstract
Chlorella vulgaris ITBBA3-12 has a role in the purification of the rubber processing wastewater. Its complete chloroplast genome contains 168369 bp, with a G + C content of 33.0%. A total of 147 genes were annotated, including 113 protein-coding genes, three rRNA (rrn23, rrn16, and rrn5) genes, and 31 tRNA genes. The significant feature of the chloroplast genome is that the genes encoding subunit V (petG), VI (petL), and apocytochrome f (petA) of the cytochrome b6/f complex are in triplicate, which was not observed in the other C. vulgaris strains. Phylogenetic analysis using the chloroplast genomes of Chlorophyta species indicated that ITBBA3-12 is closely related to C. vulgaris strain UTEX259 and NJ-7, and they clustered in the Chlorella lineage.
The Chlorella vulgaris strain ITBBA3-12 was isolated from a wastewater pond of rubber processing located in Danzhou city, Hainan Province, China with geospatial coordinates N19°30′59″, E109°29′43″. It is stored at the ClonBank of Institute of Tropical Bioscience and Biotechnology at −80 °C in 15% glycerol. Its mitochondrion genome has been reported previously (Hu et al. Citation2020). This strain is tolerate to relatively dark environment in the blackish water, therefore, its chloroplast genome is of great interest.
The genomic DNA was isolated as previously described (Ma et al. Citation2020; Yu et al. Citation2020), and sequenced with both PacBio RSII and Illumina Hiseq 2500 platforms at Genoseq (Wuhan, China). The chloroplast genome was assembled and corrected using CANU (Koren et al. Citation2017) and GATK (Zhu et al. Citation2015), respectively, and deposited in the Genome Warehouse in National Genomics Data Center, Chinese Academy of Sciences, under accession number GWHANRG00000000, and in GenBank under accession number MT920676.
The circular chloroplast genome has a length of 168,369 bp, bigger as compared to the chloroplast genomes of C. vulgaris strain C-27 (150,613 bp, MK948100) and strain NJ-7 (154,201 bp, NC_045362). The GC content is 33.0%, higher than the two sister strains (31.6%). A total of 147 genes were annotated, including 113 protein-coding genes, three rRNA (rrn23, rrn16, and rrn5) genes, and 31 tRNA genes. The protein-coding genes are mainly involved in photosynthesis (rbcL, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbL, psbN, psbT, psbZ, chlB, chlI, chlL, chlN, atpA, atpB, atpE, atpF, atpH, atpI, ycf3, ycf4, ycf62, 2 ycf78, and accD), protein synthesis (rpl12, rpl14, rpl16, rpl19, rpl2, rpl20, rpl23, rpl36, rpl5, rps11, rps12, rps14, rps18, rps19, rps2, rps3, rps4, rps7, rps8, rps9, and infA), RNA synthesis (rpoA, rpoB, rpoC1, and rpoC2), cytochrome related (3 petA, 3 petL, 3 petG, petB, petD, and ccsA), sulfate transport (cysA and cysT), hydrolyzes (I-CvuI and clpP), cell division (ftsH), and cell envelop (cemA). 30 hypothetical protein-coding genes were also identified. The 31 tRNA genes cover the transfer of all 20 amino acids, in which five are tRNA-Leu genes. The significant feature of the chloroplast genome is that the genes encoding the apocytochrome f (petA), subunit V (petG), and subunit VI (petL) of the cytochrome b6/f complex are in triplicate, which was not observed in the other C. vulgaris strains. The cytochrome b6/f complex functions in connecting photosystem II and photosystem I, and plays essential roles in regulating the electron transport (Boekema et al. Citation1994). The triplication of the cytochrome b6/f complex subunit genes may explain the wastewater tolerance in the strain ITBBA3-12.
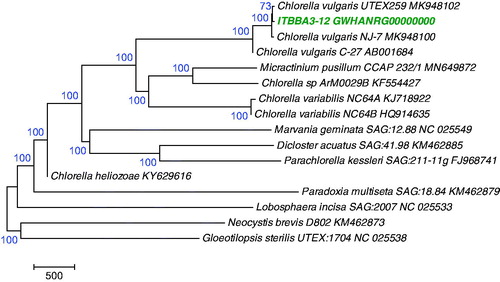
Phylogenetic analysis using the chloroplast genomes of Chlorophyta species indicated that strain ITBBA3-12 is closely related to C. vulgaris strain UTEX259 and NJ-7, and they clustered in the Chlorella lineage with 100% bootstrap support ().
Figure 1. A Maximum Likelihood tree of Trebouxiophyceae species based on chloroplast genomes. The tree was conducted in MEGA7 (Kumar et al. Citation2016) and was rooted with a representative species from Chlorophyceae (KM462873) and a species from Ulvophyceae (NC_025538), and was bootstrap-tested.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The whole genome sequence data reported in this paper has been deposited in the Genome Warehouse in National Genomics Data Center (Members Citation2019), Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under accession number GWHANRG00000000 that is publicly accessible at https://bigd.big.ac.cn/gwh. It is also available in GenBank (MT920676).
Additional information
Funding
References
- Boekema EJ, Boonstra AF, Dekker JP, Rogner M. 1994. Electron microscopic structural analysis of Photosystem I, Photosystem II, and the cytochrome b6/f complex from green plants and cyanobacteria. J Bioenerg Biomembr. 26(1):17–29.
- Hu X, Tan D, Fu L, Sun X, Zhang J. 2020. Characterization of the mitochondrion genome of a Chlorella vulgaris strain isolated from rubber processing wastewater. Mitochondrial DNA Part B. 5(3):2732–2733.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ma S, Yu B, Han B, Tan D, Fu L, Mu Y, Sun X, Zhang J. 2020. The mitochondrion genome of Heveochlorella roystonensis (Trebouxiophyceae) contains large direct repeats. Mitochondrial DNA Part B. 5(2):1355–1356.
- Members BDC. 2019. Database resources of the BIG data center in 2019. Nucleic Acids Res. 47:D8–D14.
- Yu B, Ma S, Han B, Fu L, Tan D, Sun X, Zhang J. 2020. The complete mitochondrial genome of the rubber tree endophytic alga Heveochlorella hainangensis. Mitochondrial DNA Part B. 5(2):1303–1304.
- Zhu P, He L, Li Y, Huang W, Xi F, Lin L, Zhi Q, Zhang W, Tang YT, Geng C, et al. 2015. Correction: OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PloS One. 10(9):e0138824.
