Abstract
The complete mitochondrial genome of Pontia edusa was sequenced and analyzed in the study. The length of the complete mitogenome is 15,125 bp, including 37 genes and a control region. Twelve genes start with typical ATN, but COI gene initiate with a CGA codon. Twelve of 13 PCGs have a complete stop codon TAA or TAG except for COI has an incomplete stop codon T––. Twenty-one of the 22 tRNAs have a typical clover-leaf secondary structure. The tRNASer(AGN) gene lacked the DHU loop. Bayesian analyses highly support the monophyly of Pieridae. In Pieridae, P. edusa is subordinate to the Pierinae clade.
Pontia edusa (Fabricius, 1777) is a member of the Pontia Fabricius of the subfamily Pierinae (Lepidoptera: Pieridae). The larva of P. edusa mainly harms Reseda, Turritis, Sisymbrium, Sinapis and Alyssums plants, such as Reseda odorata, Turritis glabra. The species is distributed in Europe, northwestern India, Siberia, North Africa to Ethiopia and most regions of China (Wu Citation2001). In this study, the complete mitogenome sequence of P. edusa was determined and described. The genome assembly is useful for broader research in the field of molecular ecology, systematics and phylogenetics. Meanwhile, the study also provided valuable genetic information for the identification and classification of the species more correctly. The Voucher specimen number is YF20160816 and the sequence data was deposited in GenBank under accession no. MK252290. This genome resource would be used by Shanxi Key Laboratory of Integrated Pest Management in Agriculture and Shanxi Insect Herbarium.
The adult specimen of P. edusa was collected from ShuoZhou City, Shanxi Province, China (39.45 N, 112.36E) in July 2016. Some specimens were preserved by spreading the wings in the Shanxi Insect Herbarium, College of Plant Protection, Shanxi Agricultural University and their numbers are YF20160817-20160827. The other specimens were conserved in anhydrous ethanol at −20 °C in the laboratory. Total genomic DNA was extracted from thoracic muscle of a single adult using the OMEGA Insect DNA Kit. An Illumina TruSeq library was prepared with an insert size of 350 bp and sequenced using the Illumina NovaSeq 6000 platform with 150 bp paired-end reads. Raw reads were checked by FastQC 0.11.3 (Andrews Citation2010), and low-quality reads were filtered by Trimmomatic (Bolger et al. Citation2014) and Prinseq (Schmieder and Edwards Citation2011), respectively. A total of 6 Gb clean data were obtained and assembled by IDBA-UD (Peng et al. Citation2012), and then the mitochondrial genome sequences were identified through the Geneious 10.1.3 (http://www.geneious.com/). The PCGs and two rRNA genes were confirmed by alignment with homologous genes from closely related species. The 22 tRNA genes were annotated using tRNAscan-SE version 2.0.2 (Lowe and Chan Citation2016).
The complete mitochondrial genome of P. edusa is 15,125 bp in size, containing 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. Twenty-three genes are encoded on major strand (J-strand), the other 14 genes are found in the minor strand (N-strand). The gene order and orientation are identical to the sequenced lepidopteran mitogenomes (Cao et al. Citation2013; Wang et al. Citation2013, Citation2016).
The P. edusa mitogenome is biased toward AT content at 79.91%. The nucleotide compositions are 39.91% of A, 40.00% of T, 12.23% of C, 7.85% of G. Twelve genes start with typical ATN (five with ATG, two with ATA, two with ATC, three with ATT), but COI gene initiate with a CGA codon which was observed in most lepidopteran insects (Kim et al. Citation2014; Song et al. Citation2016; Zhang et al. Citation2019). Twelve of 13 PCGs harbor the usual complete stop codon TAA or TAG, and the remaining COII gene possess the incomplete stop codons T.
The P. edusa mitogenome has the typical 22 tRNA set. The 22 tRNAs were interspersed throughout the whole mitogenome and ranged from 60 to 71 nucleotides. Twenty-one of the 22 tRNAs have a typical clover-leaf secondary structure. The tRNASer(AGN) gene lacked the DHU loop. This incomplete tRNASer (60 bp) structure has been found in many lepidopteran insects (Hong et al. Citation2009; Zhang et al. Citation2012). The rrnL is located between tRNALeu(CUN) and tRNAVal, the lengths is 1317 bp and the A + T content is 84.05%. The rrnS is located between tRNAVal and the control region, the lengths is 774 bp and the A + T content is 85.15%. The control region is located between rrnS and tRNAMet and is 373 bp long with a significant AT bias (93.03%).
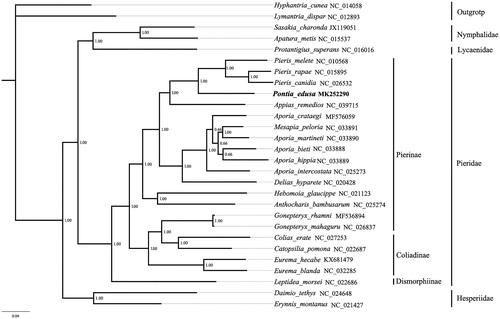
The 13 protein-coding genes of 21 known Pieridae and the other 4 Lepidoptera taxa were concatenated to reconstruct the phylogenetic relationships by Bayesian analysis (). The sequences were aligned with the Mega 7.0 software (Kumar et al. Citation2016). The aligned datasets were analyzed with the Markov Chain Monte Carlo (MCMC) algorithm under GTR + I + G model using MrBayes3.1.2 (Huelsenbeck and Ronquist Citation2001). The Hyphantria cunea (NC_014058) and Lymantria dispar (NC_012893) were selected as outgroups. Bayesian analyses highly support the monophyly of Pieridae (PP =1.00). In Pieridae, the relationship of the subfamilies is: Dismorphiinae+ (Pierinae + Coliadinae). The newly sequenced species P. edusa is subordinate to the Pierinae clade.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MK252290. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA664249, SRR12806058, and SAMN16205026 respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Cao TW, Wang JP, Xuan SB, Zhang M, Guo YP, Ma EB. 2013. Analysis of complete mitochondrial genome of Timelaea maculata (Lepidoptera, Nymphalidae). Acta Zootaxonom Sin. 38(3):468–475.
- Hong GY, Jiang ST, Yu M, Yang Y, Li F, Xue FS, Wei ZJ. 2009. The complete nucleotide sequence of the mitochondrial genome of the cabbage butterfly, Artogeia melete (Lepidoptera: Pieridae)). Acta Biochim Biophys Sin. 41(6):446–455.
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Kim MJ, Wang AR, Park JS, Kim I. 2014. Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene. 549(1):97–112.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33 (7):1870–1874.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27 (6):863–864.
- Song F, Cao TW, Cao LM, Li H, Wang JP, Xuan SB. 2016. The mitochondrial genome of the butterfly Polyura schreiber (Lepidoptera: Nymphalidae). Mitochondrial DNA Part A. 27(5):3691–3692.
- Wang JP, Cao TW, Xuan SB, Wang H, Zhang M, Ma EB. 2013. The complete mitochondrial genome of Sasakia funebris (Leech) (Lepidoptera: Nymphalidae) and comparison with other Apaturinae insects. Gene. 526(2):336–343.
- Wang JP, Xuan SB, Cao LM, Hao JS, Cao TW. 2016. The complete mitochondrial genome of the butterfly Herona marathus (Lepidoptera: Nymphalidae). Mitochondrial DNA Part A. 27(6):4399–4400.
- Wu CS. 2001. Fauna sinica insect. Vol. 52 Lepidoptera Pieridae. Beijing: Science Press.
- Zhang M, Nie X, Cao T, Wang J, Li T, Zhang X, Guo Y, Ma E, Zhong Y. 2012. The complete mitochondrial genome of the butterfly Apatura metis (Lepidoptera: Nymphalidae). Mol Biol Rep. 39(6):6529–6536.
- Zhang M, Yin J, Ma PJ, Li T, Cao TW, Zhong Y. 2019. The complete mitochondrial genomes of Aporia crataegi, Gonepteryx rhamni, and Appias remedios (Lepidoptera, Pieridae) and phylogenetic relationship of other Pieridae species. Int J Boil Macromol. 129:1069–1080.