Abstract
Cymbidium hookerianum Rchb.f. is an ornamental orchid with large flowers and delicate aroma. Here, we reported the complete chloroplast genome sequence of C. hookerianum. The total chloroplast genome cycle was 155,447 bp. It displayed a typical structure including one large single-copy (LSC, 84,186 bp) region, one small single-copy (SSC, 17,839 bp) region, and two inverted repeat (IRs, 26,711 bp). 124 genes (78 CDSs, 38 tRNAs, 8 rRNAs) were encoded by the cp genome. The average GC content of this sequence was 36.8%. The phylogenetic analysis revealed C. hookerianum and C. changningense are sisters. The groundwork of chloroplast genome would provide available reference for molecular taxonomy and breeding.
Orchidaceae is the largest family in monocotyledons. As a member of the Subgen Cyperorchis in Orchidaceae, Cymbidium hookerianum Rchb.f. is characterized by large flowers and slight aroma (Liu et al. Citation2009). Cymbidium hookerianum enjoys a shady and moist forest environment at an altitude of 1500–2600 m in northern Yunnan, eastern Nepal and north-eastern India (DuPuy et al. Citation2007). It is usually hybridized under natural conditions. Simultaneously, owing to the fascinating floral characteristics, this species has been selected as superior parents and bred hundreds of cultivars in intraspecific and interspecific (DuPuy et al. Citation1985). What is more, Cymbidum hybrids, cultivated by C. hookerianum, has become a popular production for trade worldwide. However, there are many kinds of natural and cultivated hybrids which brings a lot of difficulties in the identification and selection of parents. And most of the classification of Cymbidium is built on morphology (Yang et al. Citation2013). As for C. hookerianum, no complete genome has been reported. Now, the complete chloroplast genome helps to reveal the genetic taxonomy at molecular level, and serves as a systematic understanding of the status of this species in the genus. Also, it displays a clear guideline for breeding new cultivars.
Leaf samples of C. hookerianum were acquired from Fuzhou, Fujian province, China, and preserved in Fujian Agriculture and Forestry University (Voucher specimen: HTL-FJ2019-28A, 26°20′21.3″ N, 113°12′39.6″ E). Total genomic DNA was extracted from fresh leaves by modified CTAB procedure, and sequenced on the BGI-500 platform (BGI, Wuhan, China) (Mak et al. Citation2017). The reads length of raw sequencing data is 150 bp. About 10 Gb clean reads were obtained after moving adapters and unreliable reads by FASTQ software (Chen et al. Citation2018). Filtered data was assembled by bowtie2 and Spades (k-mer parameters: -k 21,45,65,85,105). Then, geneious prime 2020.2.1and Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) and MEGAX version10.1.8 (Kumar et al. Citation2018) were tools to accomplish annotation.
A complete circular chloroplast sequence of C. hookerianum(MT800927), with a total length of 1,55,447 bp, was established to include one large single copy (LSC, 84,186 bp) region, one small single copy (SSC,17,839 bp) region, and two inverted repeat (IRs, 26,711 bp) regions. The average GC content of this sequence was 36.8%, and every region had uneven GC content: LSC: 34.3%, SSC: 29.5%, IRs: 43.1%, respectively. It had been found that 124 genes (78 CDSs, 38 tRNAs, 8 rRNAs) were encoded in the cp genome.
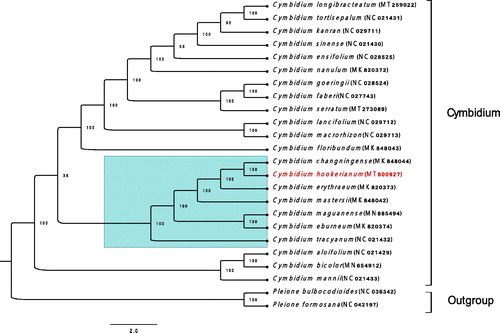
To investigate the phylogenetic relationships of C. hookerianum, the complete chloroplast sequence of 24 species, among which 22 sequences are Cymbidium and the other two are outgroups, was illustrated as the phylogenetic dendrogram. These sequences were aligned in geneious prime 2020.2.1 by Mafft Multiple Alignment plugin version 1.4.0, and a maximum-likelihood tree (ML) with 1000 bootstrap replicates was built by RAxML-HPC2 program (Stamatakis et al. Citation2008) on CIPRES (Miller et al. Citation2010). The dendrogram showed that C. hookerianum was more closely related to C. changningense and formed a clade with other 6 species from Sect. Iridorchis and Sect. Eburnea. Among the 6 species, C. tracyanum, C. erythraeum, and C. hookerianum are in the Sect. Iridorchis of Cymbidium (DuPuy et al. Citation1985). However, we are interested to notice that C. tracyanum has a further kinship with other members of Sect. Iridorchis than with Sect. Eburnea (). We believe this research gives a more valid molecular taxon evidence, and supplies effective genomic resources for Cymbidium hybridizing.
Disclosure statement
No conflict of interest was reported by the author(s).
Data availability statement
The Cymbidium hookerianum raw data has been stored in omics database of Genome Sequence Archive. GSA accession number is CRA003135. All the information can be found on the website (https://bigd.big.ac.cn/gsa/browse/CRA003135/CRX151933).
The data that support the findings will be available in GenBank at (https://www.ncbi.nlm.nih.gov/nuccore/MT800927).
Additional information
Funding
References
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- DuPuy D, Cribb P, Tibbs M. 2007. The genus Cymbidium. Kew: Royal Botanic Gardens.
- DuPuy DJ, Ford-Lloyd BV, Cribb PJ. 1985. A numerical taxonomic analysis of Cymbidium, Section Iridorchis (Orchidaceae). Kew Bull. 40(2):421.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu ZJ, Chen SC, Cribb PJ. 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong D, editors. Flora of China, vol. 25. Beijing: Science Press, Missouri Botanical Garden Press.
- Mak SST, Gopalakrishnan S, Caroe C, Geng C, Liu S, Sinding MS, Kuderna LFK, Zhang W, Fu S, Vieira FG, et al. 2017. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience. 6(8):1–13.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA. pp. 1–8.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57(5):758–771.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang J-B, Tang M, Li H-T, Zhang Z-R, Li D-Z. 2013. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 13(1):84.