Abstract
The first complete chloroplast genome of Morella cerifera was obtained by illumina platform sequencing technology in this study. The size of genome is 158,943 base pairs, consist of a pair of IRs 26,043 bp in length, the LSC region of 88,167 bp and SSC region of 18,690 bp. The genome has 112 unique genes, among which 79 protein-coding genes, 29 tRNAs, and 4 rRNAs. Phylogenetic analysis revealed that M. cerifera clustered with M. rubra within Myricaceae.
Introduction
Morella cerifera (L.) Small is the species of the genus Morella within the family of Myricaceae (Chen et al. Citation2004). M. cerifera is an evergreen dioecious plant, symbiotic with nitrogen-fixing bacteria (Frankia SP) and can grow on relatively barren sandy and alkaline soils (Zhizhen Citation2003; Karumuna et al. Citation2019). M. cerifera is one of the pioneers in afforestation improvement in saline and alkaline lands. It has the medicinal value of analgesia and treatment of digestive problems (Zhang et al. Citation2016; Makule et al. Citation2018). Chloroplast, plant-specific organelles with special genetic material, named chloroplast genome (Meng et al. Citation2018), is the site of plant photosynthesis, which can provide energy for plant synthesis of starch, amino acids and other substances (Neuhaus and Emes Citation2000). The number and sequence of genes in chloroplast genome are relatively conservative, and there is good collinearity among various plant groups (Huang et al. Citation2014; Lim et al. Citation2018; Wang et al. Citation2019). Chloroplast genome, due to its size variations, gene and intron losses, nucleotide substitutions, independent encoding specific proteins, non-recombinant and uniparental inheritance, usually has been used as ideal research model to study plant evolution and comparative genome (Liu et al. Citation2017). However, chloroplast genome of M. cerifera has never been reported. In this study, the chloroplast genome of M. cerifera was sequenced and constructed the phylogenetic analysis with other chloroplast genomes.
Plant materials were taken from Suzhou, China, located at 120.40E, 31.05 N. Voucher specimens (numbered as Morella cerifera-WF01) were collected from the Germplasm bank and were deposited at the Herbarium of Suzhou Polytechnic Institute of Agriculture (Herbarium Code: MCEM). Chloroplast DNA (cpDNA) was extracted from fresh leaves by the CTAB method (Doyle Citation1987), and the cpDNA was preserved after extraction (in Suzhou Polytechnic Institute of Agriculture). Genomic library was prepared with an insert size of 250 bp and the whole genome of M. cerifera chloroplast was sequenced using illumina platform sequencing technology (illumina NovaSeq 6000). About 5.7 G of raw reads were obtained and were filtered by the program Trimmomatic v.0.33 (Bolger et al. Citation2014). The chloroplast genome was assembled by using the filtered reads with the program Velvet 1.2.10 (Zerbino and Birney Citation2008). Gene was annotated online by GeSeq-Annotation of Organellar Genomes (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017) and the annotated genome sequence was submitted to NCBI GenBank (Accession number MT872488). The analysis results showed that the chloroplast genome length of M. cerifera is 158,943 bp, comprising a pair of IRs 26,043 bp, separating the LSC region of 88,167 bp and SSC region of 18,690 bp. The nucleotide content was 36.17%(GC) and 63.83% (AT), while the IR region has a GC content of 42.58%, significantly higher than that of the LSC and SSC regions, which are 33.87% and 29.17%, respectively.
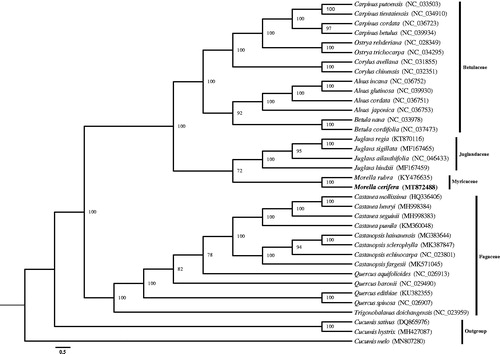
There were 112 unique genes annotated in the whole chloroplast genome of M. cerifera, including 79 protein-coding genes, 4 rRNA genes and 29 tRNA genes. Five protein-coding genes (rps7, ndhB, ycf2, rpl23, rpl2), seven tRNA genes (trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA, trnM-CAU), and four rRNA genes were duplicated within the IRs. In addition, there were 16 genes containing introns, including 15 genes containing one intron and one gene containing two introns (clpP). Thirty-five chloroplast genomes were downloaded from NCBI, of which 32 fagale chloroplast genomes and 3 cucurbitale genomes were taken as outgroups to assess the phylogenetic position of M. cerifera. Phylogenetic tree was established by using the IQ-TREE v1.6.5 with the Maximum Likelihood (ML) method (Nguyen et al., Citation2015). The tree showed a relationship that M. cerifera clustered with Morella rubra in a unique clade in family Myricaceae (). This study provides the phylogenetic status of M. cerifera with chloroplast genomic resources.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT872488. The raw sequencing reads used in this study have already been deposited in a public repository (SRA) at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA660127.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chen K, Xu C, Zhang B, Ferguson IB. 2004. Red bayberry: botany and horticulture. Oxford: John Wiley & Sons, Ltd.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang H, Shi C, Liu Y, Mao SY, Gao LZ. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151.
- Karumuna JJ, Yan D, Kyalo CM, Li Z. 2019. The complete chloroplast genome sequence of Morella salicifolia (Myricaceae): characterization and phylogenetic analysis. Mitochondrial DNA. Part B. Resources. 4(1):963–964.
- Lim CE, Lee S, So S, Han S, Choi J, Lee B. 2018. The complete chloroplast genome sequence of Asarum sieboldii Miq. (Aristolochiaceae), a medicinal plant in korea. Mitochondrial DNA. Part B. Resources. 3(1):118–119.
- Liu L, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu C. 2017. The complete chloroplast genome of chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Makule EE, Kraus B, Jürgenliemk G, Heilmann J, Wiesneth S. 2018. Dioic acid glycosides, tannins and methylated ellagic acid glycosides from Morella salicifolia bark. Phytochem Lett. 28:76–83.
- Meng J, Li X, Li H, Yang J, Wang H, He J. 2018. Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species. Molecules. 23(5):1015.
- Neuhaus HE, Emes MJ. 2000. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 51(51):111–140.
- Nguyen L-T, Schmidt H A, Von Haeseler A, Minh B Q. 2015. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution. 32(1):268–274. doi:10.1093/molbev/msu300.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang Y, Yuan X, Zhang J. 2019. The complete chloroplast genome sequence of Pometia tomentosa. Mitochondrial DNA. Part B. Resources. 4(2):3950–3951.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 18(5):821–829.
- Zhang J, Yamada S, Ogihara E, Kurita M, Banno N, Qu W, Feng F, Akihisa T. 2016. Biological activities of triterpenoids and phenolic compounds from Myrica cerifera Bark. Chem Biodivers. 13(11):1601–1609.
- Zhizhen L. 2003. The biological characteristics of Actinomycetes Frankia living in roots of Myrica rubra. Scientia Silvae Sinicae. 45(1):81–87.