Abstract
The common sunstar, Crossaster papposus, belongs to the family Solasteridae whose ordinal classification has been unstable. Here, for the first time, we assembled and annotated the complete mitochondrial genome of the common sunstar, C. papposus Linnaeus, 1767. The circular genome of C. papposus is 16,335 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, a control region, and large and small ribosomal subunits. The overall genomic structure and gene arrangement were identical to the reported mitochondrial genomes of sea star species, and a phylogenetic analysis of 13 PCGs recovers a closest relationship with the derived cluster of the paraphyletic order Valvatida.
The common sunstar, Crossaster papposus (Linnaeus 1767) is a conspicuous and ubiquitous starfish in the North Atlantic. C. papposus has a wide circumboreal distribution in all northern seas from the coastal region to oceanic depths (Clark and Downey Citation1992). The genus Crossaster belongs to the large family Solasteridae. Since Blake (Citation1987) reinstated the order Velatida, the Solasteridae has long been regarded as a member of the order Velatida. However, based on the valvatacean phylogeny acquired using three genes (12S, 16S, and early-stage histone H3), Mah and Foltz (Citation2011) assigned the Solasteridae to the order Valvatida. In the recent phylogenetic analysis of seven asteroid orders (Linchangco et al. Citation2017), the Valvatida appeared to be paraphyletic, and the members of the Solasteridae were not included in the tree. Here we present the complete mitogenome of C. papposus, which will be the first case within the Solasteridae. The whole mitogenome of Crossaster species will be useful to understand the phylogenetic context of the Solasteridae within the Asteroidea, as well as their diversity, taxonomy, and geographic distribution.
A specimen of C. papposus was collected in 2017 from the Beaufort Sea (82°46′1.7′′N, 42°32′52.4′′W) using a remotely operated underwater vehicle (ROV) of Monterey Bay Aquarium Research Institute (MBARI). The voucher specimen was registered both in the Research Institute of Basic Sciences of Incheon National University and in the Korea Polar Research Institute (Species ID: Echinodermata-01; Specimen ID: KOPRI-Benthos-01). Mitochondrial genomes were recovered by de novo assembling from Illumina shotgun sequence data. Genomic DNA was isolated using a QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany). Based on the manufacturer’s instructions (Illumina, San Diego, CA), a genomic library was constructed using a TruSeq Nano DNA Kit by Macrogen, Inc. (Seoul, South Korea). Raw reads were obtained from the sample that passed quality control by Illumina HiSeq platform. After the trimming process on the raw reads, de novo assembly was performed using SPAdes version 3.11.1 (Bankevich et al. Citation2012). Genomic features and annotations were performed using MITOS2 (Bernt et al. Citation2013) and tRNAscan-SE 2.0 (Lowe and Eddy Citation1997). The annotated gene structure was further confirmed using NCBI-BLAST (http://blast.ncbi.nlm.nih.gov). Mitochondrial genome of C. papposus was aligned with mitogenomes from other echinoderm genera, as well as two outgroup taxa from the genus Balanoglossus, and 13 PCGs were extracted for phylogenetic analysis (Nam et al. Citation2020). jModelTest version 2.1.10 (Darriba et al. Citation2012) was used to determine the best substitution model with an appropriate partitioning scheme, and a maximum likelihood phylogenetic analysis was conducted with 1000 bootstrap replicates in the PhyML version 2.4.5 (Guindon and Gascuel Citation2003).
The complete mitochondrial genome for C. papposus contained 13 PCGs, 22 tRNA genes, 2 rRNA genes, and a control region. The mitogenome for C. papposus (GenBank accession no. MW046047) was 16,335 bp long and had a GC content of 32.8% with an AT bias (A: 35.5%; T: 31.7%; G: 12.7%; C: 20.1%). The arrangement of genes and gene composition were identical to those of other starfishes. Our COI sequence was identical to the partial COI sequence (841 bp; GenBank accession no. MK270384) of C. papposus collected from the Baffin Bay (Ringvold and Moum Citation2020).
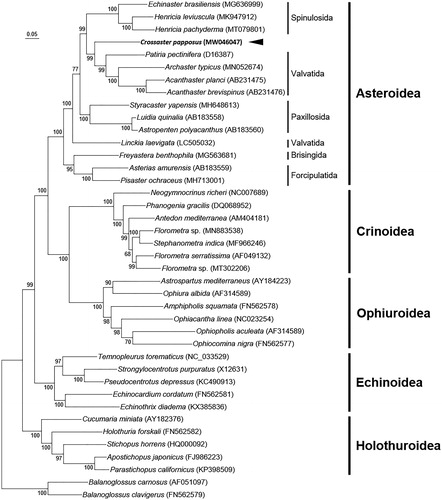
Although data for the order Velatida is lacking, our phylogenetic analysis using mitogenome data shows a similar ordinal relationship within the Asteroidea to the result of Linchangco et al. (Citation2017). Our result resolved a close relationship of C. papposus to the derived cluster of the paraphyletic Valvatida, together which form a sister group to the order Spinulosida (). This is in line with the previous suggestion that the Solasteridae belongs to the order Valvatida (Mah and Foltz Citation2011). Although different taxa of the Valvatida were used for phylogenic analysis, paraphyly of the order Valvatida was produced both in our study and in Linchangco et al. (Citation2017). Future researches on the paraphyly of the Valvatida and the mitogenome data from the order Velatida are required.
Figure 1. Maximum-likelihood (ML) phylogeny of 38 echinoderms (15 asteroids including C. papposus, 7 crinoids, 5 echinoids, 5 holothuroids, and 6 ophiuroids) and two Balanoglossus mitogenomes as an outgroup based on the concatenated nucleotide sequences of entire protein-coding genes (PCGs). Numbers at nodes represent ML bootstrap percentages (1000 replicates). DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated. The black arrow indicates the C. papposus analyzed in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MW046047.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Blake DB. 1987. A classification and phylogeny of post-Palaeozoic sea stars (Asteroidea: Echinodermata). J Nat Hist. 21(2):481–528.
- Clark AM, Downey ME. 1992. Starfishes of the Atlantic. London: Chapman & Hall.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704.
- Linchangco GV, Foltz DW, Reid R, Williams J, Nodzak C, Kerr AM, Miller AK, Hunter R, Wilson NG, Nielsen WJ, et al. 2017. The phylogeny of extant starfish (Asteroidea: Echinodermata) including Xyloplax, based on comparative transcriptomics. Mol Phylogenet Evol. 115:161–170.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Mah C, Foltz D. 2011. Molecular phylogeny of Valvatacea (Asteroidea: Echinodermata). Zool J Linn Soc. 161(4):769–788.
- Nam SE, Park HS, Rhee JS. 2020. Characterization and phylogenetic analysis of the complete mitochondrial genome of Florometra species (Echinodermata, Crinoidea). Mitochondrial DNA. 5(2):2010–2011.
- Ringvold H, Moum T. 2020. On the genus Crossaster (Echinodermata: Asteroidea) and its distribution. PLOS One. 15(1):e0227223
