Abstract
The red-necked longhorn beetle Aromia bungii is a major pest of peach orchards. In this study, we sequenced and analyzed the complete mitochondrial genome (mitogenome) of A. bungi,i. This mitogenome was 15,760 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs) and two ribosomal RNA unit genes (rRNAs). Gene order was conserved and identical to most other previously sequenced Cerambycidae. Most PCGs of A. bungii have the conventional start codons ATN (six ATT, five ATG and one ATC), with the exception of nad1 (TTG). Except for three genes (cox1, cox2 and nad5) end with the incomplete stop codon T−, all other PCGs terminated with the stop codon TAA or TAG. The whole mitogenome exhibited heavy AT nucleotide bias (74.3%). Phylogenetic analysis positioned A. bungii in a well-supported clade within the subfamily Cerambycinae with Xystrocera globosa (tribe Xystrocerini). These results support the currently accepted taxonomy and provide a better understanding of the phylogenetic analysis of the Cerambycidae.
Aromia bungii (Faldermann) (Coleoptera: Cerambycidae) is a wood-boring pest of trees in the Rosaceae family, particularly peach, cherry, plum, apricot and pear (Men et al. Citation2019). This beetle is native to China, Mongolia, Korean Peninsula, Vietnam, Russia, and has been introduced into Germany, Italy, and Japan since 2011 (Fukaya et al. Citation2017). As A. bungii spends most of its life in a larval stage under the bark, chemical control with pesticides is labor-intensive, costly, and difficult. To get more information about this species, we sequenced and analyzed its mitogenome in this study.
Specimens of A. bungii were collected from Jingangshan City, Jiangxi Province, China (26°34′N, 114°05′E, July 2019) and were stored in Entomological Museum of Gannan Normal University (Accession number GNU-EAB09). Total genomic DNA was extracted from tissues using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany). The mitogenome sequence of A. bungii was generated using Illumina HiSeq 2500 Sequencing System (Illumina, San Diego, CA). A total of 32.8 million reads were generated and had been deposited in the NCBI Sequence Read Archive (SRA) with accession number SRR12785341. Then, raw reads were assembled using MITObim v 1.7 (Hahn et al. Citation2013). By comparison with the homologous sequences of other Cerambycidae species from GenBank, the mitogenome of A. bungii was annotated using software GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand).
The complete mitogenome of A. bungii is 15,760 bp in length (GenBank accession no. MT371041), and containing the typical set of 13 protein-coding, two rRNA and 22 tRNA genes, and one non-coding AT-rich region. Gene order was conserved and identical to most other previously sequenced Cerambycidae (Li et al. Citation2016; Liu et al. Citation2018; Behere et al. Citation2019; Wang et al. Citation2019). The nucleotide composition of the mitogenome was biased toward A and T, with 74.3% of A + T content (A 39.8%, T 34.5%, C 14.8%, G 10.9%). Four PCGs (nad4, nad4l, nad5 and nad1) were encoded by the minority strand (N-strand) while the other nine were located on the majority strand (J-strand). Most PCGs of A. bungii have the conventional start codons ATN (six ATT, five ATG and one ATC), with the exception of nad1 (TTG). Except for three genes (cox1, cox2 and nad5) end with the incomplete stop codon T−, all other PCGs terminated with the stop codon TAA or TAG. The 22 tRNA genes vary from 63 bp (trnE) to 71 bp (trnK). Two rRNA genes (rrnL and rrnS) locate at trnL1/trnV and trnV/control region, respectively. The lengths of rrnL and rrnS in A. bungii are 1282 and 781 bp, with the AT contents of 79.5% and 76.7%, respectively.
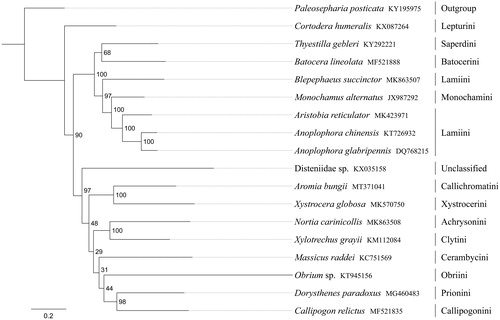
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 18 Coleoptera species. Phylogenetic tree was constructed through raxmlGUI 1.5 (Silvestro and Michalak Citation2012). Phylogenetic analysis positioned A. bungii in a well-supported clade within the subfamily Cerambycinae with Xystrocera globosa (tribe Xystrocerini) (). The relationships ((Callichromatini + Xystrocerini) + ((Achrysonini + Clytini) + (Cerambycini + (Obriini + (Callipogonini + Prionini))))) were supported in Cerambycinae, and Cerambycinae was sister to Lamiinae within Cerambycidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MT371041, SRR12785341.
Additional information
Funding
References
- Behere GT, Tay WT, Firake DM, Kunz D, Burange PS, Ramamurthy VV. 2019. Characterization of draft mitochondrial genome of guava trunk borer, Aristobia reticulator (Fabricius, 1781) (Coleoptera: Cerambycidae: Lamiinae) from India. Mitochondrial DNA Part B. 4(1):1592–1593.
- Fukaya M, Kiriyama S, Yasui H. 2017. Mate-location flight of the red-necked longicorn beetle, Aromia bungii (Coleoptera: Cerambycidae): an invasive pest lethal to Rosaceae trees. Appl Entomol Zool. 52(4):559–565.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Li W, Yang X, Qian L, An Y, Fang J. 2016. The complete mitochondrial genome of the citrus long-horned beetle, Anoplophora chinensis (Coleoptera: Cerambycidae). Mitochondrial DNA Part A. 27(6):4665–4667.
- Liu Y, Chen D, Liu H, Hu H, Bian H, Zhang R, Yang R, Jiang X, Shi S. 2018. The complete mitochondrial genome of the longhorn beetle Dorysthenes paradoxus (Coleoptera: Cerambycidae: Prionini) and the implication for the phylogenetic relationships of the Cerambycidae species. J Insect Sci. 18(2):21–29.
- Men J, Zhao B, Cao D, Wang W, Wei J. 2019. Evaluating host location in three native Sclerodermus species and their ability to cause mortality in the wood borer Aromia bungii (Coleoptera: Cerambycidae) in laboratory. Biol Control. 134:95–102.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers E. 12(4):335–337.
- Wang J, Dai X, Xu X, Zhang Z, Yu D, Storey KB, Zhang J. 2019. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ. 7(9):e7633.