Abstract
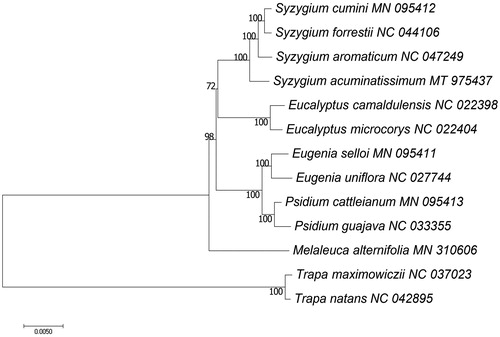
Syzygium acuminatissimum is a valuable hard wood species in southern China. In this study, we sequenced, assembled and annotated the complete chloroplast genome of S. acuminatissimum. The complete cp genome of S. acuminatissimum was 159,352 bp in length, with a total of 109 unique annotated genes, including 78 protein-coding genes, 27 tRNA genes and 4 rRNA genes. Phylogenetic analysis showed that S. acuminatissimum was closely related to its congener S. aromaticum.
Introduction
Syzygium acuminatissimum (Blume) Candolle, belonging to Myrtaceae, is an evergreen tree distributed in the low-latitude subtropical forests of China (Peng and Wang Citation1995). With perfectly straight trunk and hard wood, it is a valuable material for furniture (Zhou Citation2001). Previous authors studied leaf photoprotection and cultivation of this species (Liu et al. Citation2013; Zhu et al. Citation2018), and phylogenetic position of Syzygium in the family Myrtaceae was resolved based on sequences of three chloroplast (cp) fragments (Biffin et al. Citation2006). In this study, we assembled and annotated the complete cp genome of S. acuminatissimum, and investigated its phylogenetic relationship with related species in this family.
Fresh leaves of an S. acuminatissimum individual were collected from the Conghua Baimu Nursery (113°24′06″E, 23°43′04″N), Guangzhou and the voucher specimen (Zhang-20200729) was deposited at the Herbarium of Sun Yat-sen University (SYS), Guangzhou, China. Genomic DNA was extracted using the CTAB method (Doyle and Doyle Citation1987), and a DNA shotgun library was constructed according to the manufacturer’s protocol (reads size: 250 bp) and sequenced on an Illumina HiSeq X TEN platform (Illumina, San Diego, CA, USA). After quality control with SOAPnuke (Chen et al. Citation2018), about 2 Gb of clean reads was used to assemble the complete cp genome using SPAdes v3.13.0 (Bankevich et al. Citation2012), with the cp genome of Syzygium cumini (MN095412, template size: 1,58,499 bp) as the reference. Gene annotation was conducted by GeSeq (Tillich et al. Citation2017). The annotated cp genome sequence was submitted to GenBank (accession number: MT975437).
The complete cp genome of S. acuminatissimum was 1,59,352 bp in length with a typical quadrant structure, including a large single-copy (LSC) of 87,993 bp, a small single-copy (SSC) of 18,417 bp, and a pair of inverted repeats (IRs) of 26,471 bp. A total of 109 unique genes, including 78 protein coding genes (PCGs), 27 tRNAs and 4 rRNAs, were annotated. The overall GC content of the cp genome was 34.73%. To resolve the phylogenetic relationships of S. acuminatissimum with other species in Myrtaceae, we selected 11 species from Myrtaceae as ingroups and 2 species from Lythraceae as outgroups. Based on 78 common PCGs, we used RAxML (Stamatakis Citation2014) to construct maximum likelihood (ML) tree with 1000 bootstrap replicates. The phylogenetic tree showed that S. acuminatissimum was closely related to S. aromaticum (). The cp genome of S. acuminatissimum in this study provides useful information for future phylogenetic research on this genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data of this study are available on GenBank with the accession number MT975437 (https://www.ncbi.nlm.nih.gov/nuccore/MT975437).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Biffin E, Craven LA, Crisp MD, Gadek PA. 2006. Molecular systematics of Syzygium and allied genera (Myrtaceae): evidence from the chloroplast genome. Taxon. 55(1):79–94.
- Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. 2018. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7(1):1–6.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Liu J, Huang W, Zhou G, Zhang D, Liu S, Li Y. 2013. Nitrogen to phosphorus ratios of tree species in response to elevated carbon dioxide and nitrogen addition in subtropical forests. Glob Chang Biol. 19(1):208–216.
- Peng S, Wang B. 1995. Forest succession at Dinghushan, Guangdong, China. Chin Sci Abstracts Ser B. 14:35–36.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhou T. 2001. Cultivation techniques of main economic trees in Tropical China. Beijing: China Forestry Press. p. 292–294.
- Zhu H, Zhang T-J, Zheng J, Huang X-D, Yu Z-C, Peng C-L, Chow WS. 2018. Anthocyanins function as a light attenuator to compensate for insufficient photoprotection mediated by nonphotochemical quenching in young leaves of Acmena acuminatissima in winter. Photosynt. 56(1):445–454.