Abstract
The black-spotted stout newt (Pachytriton brevipes) is widely distributed in the mountains of southeastern China. In this study, we sequenced and determined the complete mitochondrial genome (mitogenomes) of two P. brevipes samples collected in Mount Wuyi. The assembled mitogenomes were 16,298 bp and 16,301 bp in length, and contained 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, one non-coding region, and one control region. The phylogenetic analysis indicated the two samples of black-spotted stout newt grouped together and are the sister group of P. feii.
The black-spotted stout newt (Pachytriton brevipes) is an endemic newt, which mainly inhabits mountains 800–1700 m high in southeastern China including eastern Jiangxi, Guangdong, Zhejiang, and Fujian (Frost Citation2020). Up to present, 10 species have been recorded within the genus Pachytriton, and only occur in southeastern China (Frost Citation2020). The mitochondrial genome (mitogenome), is an effective genetic marker, that has been used to provide molecular support for phylogenetic analyses. We sequenced and annotated the complete mitogenome of P. brevipes and analyzed phylogenetic relationships among other Salamandridae.
In this study, one adult assigned to P. brevipes with larger black spots (LSU20200714WY01) and another specimen with smaller black spots (LSU20200716WY01) were collected in two streams (N27.69114°, E117.65062°; N27.71714°, E117.64825°) at Dazhulan from Mount Wuyi National Park, Fujian Province, China, respectively. Both specimens were preserved in 90% ethanol and deposited at the Museum of the Laboratory of Amphibian Diversity Investigation at Lishui University. Genomic DNA was extracted from muscles using the EasyPure genomic DNA kit (TransGen Biotech Co., Beijing, China). The mitogenomes were sequencing by Illumina NovaSeq 6000 platform (Novogene Bioinformatics Technology Co. Ltd., Tianjin, China) for PE 2 × 150 bp. Raw data of two samples (31.14 G) were deposited in the NCBI’s Sequence Read Archive (SRA) database (accession: SRR12673759 and SRR12673760), and clean data without sequencing adapters were de novo assembled by the NOVO Plasty 3.7 (Dierckxsens et al. Citation2017).
The lengths of complete and circular mitogenome sequence are 16,301 bp (GenBank accession: MT993830) and 16,298 bp (GenBank accession: MW004669) in individuals with larger black spots and smaller black spots, respectively. The base composition was 28.4% T, 23.5% C, 33.7% (or 33.8%) A, and 14.4% (or 14.3%) G, demonstrating a bias of higher AT content (62.07%% and 62.15%) than GC content (37.93% and 37.85%). Although there are differences in the size of black spots between the two P. brevipes samples, the composition and arrangement of mitochondrial genes are consistent. In P. brevipes, the complete mitogenome sequence contained 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (12S rRNA and 16S rRNA) genes, one non-coding region (NCR), and one control region (CR). The positions of 37 mitochondrial genes, NC region, and CR were predicted using the MITOS Web Server (Matthias et al. Citation2013). Except eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro), and ND6, all rest genes were encoded on the heavy strand. Most PCGs start with an initiation ATG codon, except COI starts with GTG. COI, ATP6, ATP8, ND2, ND3, ND4L, and ND5 used TAA as their stop codon; ND1 used TAG as its stop codon; ND6 used AGA as its stop codon; and the rest four PCGs (COII, COIII, ND4, and Cytb) were terminated by incomplete TA(A) or T(AA). The CR is 733 bp in length and the NCR (30 bp) is found between tRNAAsn and tRNACys. The gene arrangement and composition were identical to previous studies in the genus Pachytriton (Fang et al. Citation2017).
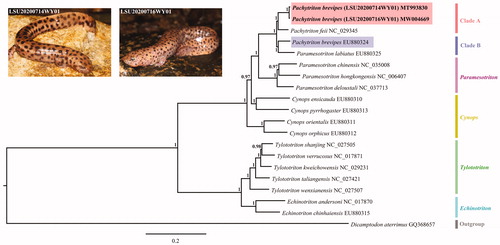
Bayesian analyses based on the 13 PCGs (11,379 bp) of 19 species of the Salamandridae family were performed with MrBayes 3.2.1 (Ronquist and Huelsenbeck Citation2003). Dicamptodon aterrimus (Caudata: Dicamptodontidae) was chosen as the outgroup. We used MrModelTest 2.3 (Nylander Citation2004) to obtain the best-fit substitution model (GTR + I+G). As shown in , our two samples of P. brevipes (clade A) grouped together and were retrieved as sister group of P. feii. The other P. brevipes sample (blue bar) clustered with Paramesotriton labiatus (clade B). However, our phylogenetic relationships within the genus Pachytriton were not similar to the previous study reported by Zhang et al. (Citation2008). According to our results, either the two genera Pachytriton and Paramesotriton are paraphyletic or the specimen Pachytriton brevipes sequenced in Zhang et al. (Citation2008) was misidentified. The new complete mitogenomes of P. brevipes from this study will contribute to the further understanding of the genus Pachytriton.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI at www.ncbi.nlm.nih.gov [MT993830 and MW004669].
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fang K, Zhang CL, Guo WB, Lifu Qian LF, Wu J, Zhang BW. 2017. Mitochondrial genome of Pachytriton feii (Urodela: Salamandridae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):288–289.
- Frost DR. 2020. Amphibian species of the world: an online reference, version 6.0. New York (NY): American Museum of Natural History; [accessed 2020 Sep 18]. http://research.amnh.org/herpetology/amphibia/index.php/.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Zhang P, Papenfuss TJ, Wake MH, Qu L, Wake DB. 2008. Phylogeny and biogeography of the family Salamandridae (Amphibia: Caudata) inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 49(2):586–597.