Abstract
Scutellaria scordifolia Fisch. ex Schrank Li is a traditional Chinese medicinal plant of genus Scutellaria from the Labiatae family. The complete chloroplast genome of was 152,336 bp in length, which contained 133 complete genes including 87 protein-coding genes (87 PCGs), 8 ribosomal RNA genes (8 rRNAs), and 37 transfer RNA genes (37 tRNAs). The GC content of chloroplast DNA was 38.3%. The corresponding values of the LSC, SSC, and IR regions were 36.3%, 32.5%, and 43.6%, respectively. Phylogenetic tree showed that the species from genus Scutellaria were divided into two monophyletic clades, and the divergence time of S. scordifolia was earlier than that of the other species.
Scutellaria scordifolia Fisch. ex Schrank Li is a traditional Chinese medicinal plant of genus Scutellaria from the Labiatae family (Bruno et al. Citation2002). This species mainly distributes in grassland and wet meadow in the provinces of Inner Mongolia, Heilongjiang, Hebei, Shanxi, and Qinghai. The dried root of S. scordifolia, as a traditional herb medicine, is used to treat jaundice, liver heat, hepatomegaly, gingival abscess, etc. in China, Japan, and South Korea. It has been reported to have antiviral, antimycotic, antioxidant, nitric-oxide-inducing, and quinone-reductase-inducing activities (Shao et al. Citation1999; Li et al. Citation2000; Ng et al. Citation2000). However, due to anthropogenic over-exploitation and decreasing distributions, this species is in urgent need of protection. Knowledge of the genetic information of this species will contribute to the protection policy for this species. In this study, we assembled and characterized the complete chloroplast (cp) genome of S. scordifolia, hoping to lay a foundation for further research.
Fresh leaves of S. scordifolia were collected in Haiyuan County of Ningxia province, China (105°38′E, 36°21′N) and dried with silica gel. The voucher specimen was stored in Sichuan University Herbarium (JQ13383018). Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987) and a 350-bp library was constructed. This library was sequenced on the Illumina NovaSeq 6000 system with 150 bp paired-end reads. We obtained 10 million high quality pair-end reads from S. scordifolia, and after removing the adapters, the remained reads were used to assemble the complete chloroplast genome by NOVOPlasty (Dierckxsens et al. Citation2017). The complete chloroplast genome of S. insignis was used as a reference. Plann v1.1 (Huang and CronK Citation2015) and Geneious v11.0.3 (Kearse et al. Citation2012) were used to annotate the chloroplasts genome and correct the annotation.
The total plastome length of S. scordifolia (MT712016) was 152,366 bp, which exhibited a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 84,407 bp, two inverted repeat (IR) regions of 25,201 bp and a small single copy (SSC) region of 17,556 bp. The cp genome contained 133 complete genes, including 87 protein-coding genes (87 PCGs), 8 ribosomal RNA genes (8 rRNAs) and 37 transfer RNA genes (37 tRNAs). Most genes occurred in a single copy, while 17 genes occurred in double, including all rRNAs (4.5S, 5S, 16S, and 23S rRNA), 7 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACC, and trnV-GAC), and 6 PCGs (rps7, ndhB, ycf2, ycf15, rpl2, and rpl23). The GC content of cp DNA was 38.3%. The corresponding values of the LSC, SSC, and IR regions were 36.3%, 32.5%, and 43.6%, respectively.
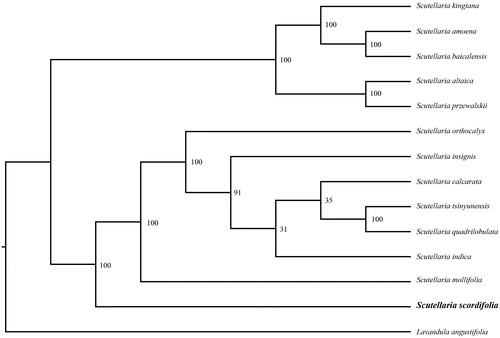
In order to further clarify the phylogenetic position of S. scordifolia, plastome of 12 representative scordifolia species were obtained from NCBI to reconstruct the plastome phylogeny with Lavandula angustifolia as an outgroup. All the sequences were aligned by MAFFT v.7.313 (Katoh and Standley Citation2013) and maximum likelihood (ML) phylogenetic analyses were conducted by RAxML v.8.2.11 (Stamatakis Citation2014) under GTRCAT model with 500 bootstrap replicates. The phylogenetic tree showed that the species of genus Scutellaria were divided into two monophyletic clades. S. kingiana, S. amoena, S. baicalensis, S. altaica, and S. przewalskii clustered together in one clade, while S. scordifolia was identified as the basal subclade of the other clade, indicating that the divergence time of S. scordifolia was earlier than that of the other species ().
Figure 1. Phylogenetic relationships of Scutellaria using complete chloroplast genome.
GenBank accession numbers: Lavandula angustifolia (NC_029370), Scutellaria insignis (NC_028533), Scutellaria indica (MN047312), Scutellaria calcarata (MN128385), Scutellaria baicalensis (MF521633), Scutellaria amoena (MN128386), Scutellaria altaica (MN128387), Scutellaria tsinyunensis (NC_050161), Scutellaria quadrilobulata (MN128381), Scutellaria przewalskii (MN128382), Scutellaria orthocalyx (MN128383), Scutellaria mollifolia (MN128384), and Scutellaria kingiana (MN128389).
Acknowledgements
We gratefully acknowledge the support of Dr. Lei Zhang for his field sampling and helpful comments. We are grateful to the opened raw genome data from public database.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov). The reference number of the data is MT712016.
Additional information
Funding
References
- Bruno M, Piozzi F, Maggio AM, Simmonds MS. 2002. Antifeedant activity of neoclerodane diterpenoids from two Sicilian species of Scutellaria. Biochem Syst Ecol. 30(8):793–799.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: A command-line application for anno-tating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li BQ, Fu T, Gong WH, Dunlop N, Kung H, Yan Y, Kang J, Wang JM. 2000. The flavonoid baicalin exhibits anti-inflammatory activity by binding to chemokines. Immunopharmacology. 49(3):295–306.
- Ng TB, Liu F, Wang ZT. 2000. Antioxidative activity of natural products from plants. Life Sci. 66(8):709–723.
- Shao ZH, Li CQ, Hoek TL, Becker LB, Schumacker PT, Wu JA, Attele AS, Yuan CS. 1999. Extract from Scutellaria baicalensis Georgi attenuates oxidant stress in cardiomyocytes. J Mol Cell Cardiol. 31(10):1885–1895.
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
