Abstract
Muscina pascuorum (Diptera: Muscidae) represents an important hygiene pest. The complete mitochondrial genome (mitogenome) of M. pascuorum was first sequenced and annotated in this study. The full length of mitogenome was 14, 940 bp, consisting of 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA), and one AT-rich region. The nucleotide content of these flies was 40.0% A, 13.2% C, 9.1% G, and 37.6% T. This study illustrates that the arrangement of the genes was identical to classical metazoans. Besides, the phylogenetic analyses indicated that the branch of M. pascuorum was clustered separately from the common three Muscina spp in the tree. This genome provides an essential reference for understanding the phylogenetic relationships of Muscidae.
The Muscidae is of great significance in hygienic pests, covering almost the whole world (Ren et al. Citation2018). Muscina pascuorum Meigen, 1826, belonging to Muscidae family and Diptera order, is usually found entirely in late summer and autumn (Johnson Citation1926). The adults tend to be live in places with cover soft fungus-like swamps, puddle, and lawn (Curran Citation1926). Compared with single genes or subregions of the mitochondrial genomes (mitogenomes), mitogenomes have remarkably improved the accuracy of taxonomic relationships and the efficacy of phylogenetic analyses, especially for closely related species (Nadimi et al. Citation2016). In this study, the full length of mitogenome was 14,940 bp (GenBank accession no. MT017715), consisting of 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA), and one AT-rich region. The nucleotide content of these flies was 40.0% A, 13.2% C, 9.1% G, and 37.6% T.
Adult specimens of M. pascuorum were captured in Ürümqi city (43°50′N, 87°37′E), Xinjiang province, China, in June 2019. These species were identified by a morphological expert and then preserved at −80 °C in Guo’s lab (Changsha, China) with a unique code (CSU19111968). The DNA was extracted from thoracic tissues of the flies by the QIANamp Micro DNA Kit (Qiagen Biotech Co., Ltd., Shanghai, China). Sequences were performed on an Illumina HiSeq 2500 Platform. Mitochondrial de novo assembly was performed using MITObim v 1.9 and SOAPdenovo v2.04 (https://github.com/chrishah/MITObim and http://soap.genomics.org.cn/soapdenovo.html). Finally, the rough boundaries of each gene were initially identified using MITOS2 Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Ren et al. Citation2020).
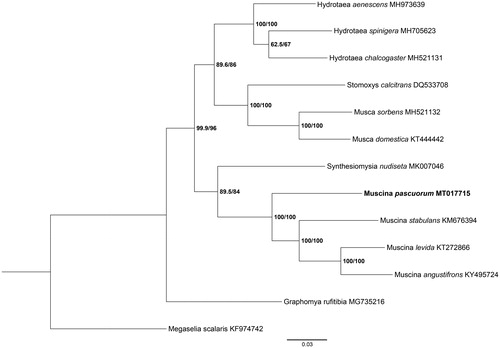
Phylogenetic analysis included M. pascuorum and 11 Muscidae species using maximum likelihood (ML) method based on 13 PCGs and Megaselia scalaris as the outgroup (). ML analysis was carried out with IQ-TREE v1.6.12 (Ren et al. Citation2020). The tree showed that M. pascuorum belongs to the Muscina subgenus, which was clearly separated from the clade of Muscidae species. This study provided several crucial genetic data of dipteran mitogenomes for further study on evolution analysis and species identification.
Acknowledgements
We are grateful to Prof. Lushi Chen (Guizhou Police College) for species identification.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The raw data that support the findings of this study are openly available in the SRA (Sequence Read Archive) database of NCBI (National Center for Biotechnology Information) via accession numbers at https://www.ncbi.nlm.nih.gov/sra/?term=SRR12587884 (SRR12587884). The assembled mitochondrial genome is available on NCBI at https://www.ncbi.nlm.nih.gov (accession No. MT017715).
Additional information
Funding
References
- Curran CH. 1926. The distribution in Canada of the European scavenger fly, Muscina Pascuorum Mg. Can Entomol. 58(10):235–236.
- Johnson CW. 1926. The distribution of Muscina pascuorum Meigen in America. Psyche J Entomol. 33(1):20–21.
- Nadimi M, Daubois L, Hijri M. 2016. Mitochondrial comparative genomics and phylogenetic signal assessment of mtDNA among arbuscular mycorrhizal fungi. Mol Phylogenet Evol. 98:74–83.
- Ren L, Chen W, Shang Y, Meng F, Zha L, Wang Y, Guo Y. 2018. The application of COI gene for species identification of forensically important muscid flies (Diptera: Muscidae). J Med Entomol. 55(5):1150–1159.
- Ren L, Zhang X, Li Y, Shang Y, Chen S, Wang S, Qu Y, Cai J, Guo Y. 2020. Comparative analysis of mitochondrial genomes among the subfamily Sarcophaginae (Diptera: Sarcophagidae) and phylogenetic implications. Int J Biol Macromol. 161:214–222.