Abstract
The silverfish Neoasterolepisma foreli belongs to the family Lepismatidae within Zygentoma and is well known for the peculiar habit of living in strict association with ant nests (myrmecophily). In this study, we describe its mitochondrial genome, a circular molecule of 15,398 bp including the canonical 13 PCGs, 22 tRNAs, 2 rRNAs, as well as a 403 bp AT-rich region. A phylomitogenomic analysis of the new sequence, alongside basal hexapod mtDNAs, confirmed the monophyly of all orders, with some uncertainty over the position of the enigmatic Tricholepidion gertschi that would make Zygentoma paraphyletic. Neoasterolepisma foreli is recovered in a basal position within family Lepismatidae, at odd with our current understanding of the group that would, in turn, suggest a closer relationship with the genus Lepisma (Mendes, Citation1991).
Zygentoma (silverfishes) is a small taxon of primitively wingless hexapods. Despite the availability of anatomical and embryological data (Gaju-Ricart et al. Citation2015), genetic and genomic information is still limited. They are generally regarded as sister group of the flying insects (e.g. Mendes Citation2018), with some doubts on the enigmatic Tricholepidion gertschi that may represent an early offshoot of the common dicondylian stock, making Zygentoma the only non-monophyletic high-ranking basal hexapod group. This latter hypothesis was proposed based on morphological evidence (i.e. Boudreaux Citation1979; but see Kristensen Citation1998; Blanke et al. Citation2014) and revived based on phylomitogenomic investigations (Carapelli et al. Citation2006; Leo et al. Citation2019). Lepismatidae are the largest family within Zygentoma, display a cosmopolitan distribution and include well known anthropophilic species such as Lepisma saccharinum and Thermobia domestica. Nevertheless, infraorder relationships are still unclear. The species Neoasterolepisma foreli (Moniez, 1894) has been investigated in relation to its myrmecophilous habit (Molero-Baltanás et al. Citation2017), but its phylogenetic position has never been studied in detail. In this work, we describe the complete mitochondrial genome of Neoasterolepisma foreli and study its phylogenetic position within basal Dicondylia.
A single specimen of N. foreli collected in 2019 in Vélez de Benaudalla (Granada, Spain; 36°50'13″N 3°30′24″W; voucher ID: NFO_03, preserved at the Life Sciences Department of the University of Siena; determined by M. G. -R.) was used for total genomic extraction using QIAmp® UCP DNA kit. Libraries were prepared using the TruSeq DNA Nano kit (Illumina, San Diego, CA) and 151 bp paired-end sequences were obtained on a HISeq 2500 platform (Illumina, San Diego, CA) at Macrogen Europe together with additional libraries from unrelated arthropod species (data not shown). Resulting reads were de novo assembled as in Nardi et al. (Citation2020). In brief, the cox3 gene was employed as seed in the NOVOPlasty v. 3.8.3 (Dierckxsens et al. Citation2017) assembling method with manually curated k = 33 and 39. The resulting assemblies were checked against a MEGAHIT assembly (Li et al. Citation2015) and manually curated. The final mtDNA sequence was automatically annotated using MITOS (Bernt et al. Citation2013) and manually curated. The new sequence (PCGs only) was retro-aligned with the dataset of Leo et al. (Citation2019), together with Ctenolepisma villosum (Chen et al. Citation2019) and Lepisma saccharinum (Bai et al. Citation2020) sequences, and hypervariable regions were removed using Gblocks v. 0.91b (Castresana Citation2000). The best partitioning scheme and evolutionary model were selected using PartitionFinder 2.1.1 (Lanfear et al. Citation2016) and used for a Bayesian phylogenetic analysis in MrBayes 3.2.6 (Ronquist et al. Citation2012) through the CIPRES Science Gateway (Miller et al. Citation2010) with four chains of 2 × 107 generations, a burn-in of 0.25 and a sampling frequency of one tree every 1000 iterations.
The complete mtDNA of N. foreli is 15,398 bp long, in line with the previously described mitogenomes of Lepismatidae (e.g. Thermobia domestica: 15,152 bp, Lepisma saccharinum: 15,244 bp, Ctenolepisma villosum: 15,488 bp). The molecule encodes for the common 37 genes encountered in metazoan mtDNAs (13 PCGs, 22 tRNAs, and 2 rRNA) plus a non-coding A + T rich region of 403 bp. Overall, all PCGs show a canonical Methionine start codon (ATN), with the exception of cox1 (TTG – Leucine), nad3 and nad5 (ATT – Isoleucine). All PCGs are characterized by a TAA complete stop codon, with the exception of nad1 (TAG), nad3 and cob (T–). Two overlaps between adjoining genes are observed, namely atp8–atp6 (4 nt) and nad4–nad4L (7 nt). Gene order conforms to the ancestral Pancrustacea model (Boore Citation1998), as in all other silverfishes studied (Comandi et al. Citation2009). The nucleotide composition of the entire mitogenome is as follows: A = 39.8%, T = 32.2%, C = 17.6% and G = 10.4%, with the highest AT nucleotide bias (72%) within the order (Cook et al. Citation2005; Comandi et al. Citation2009; Chen et al. 2019; Bai et al. Citation2020).
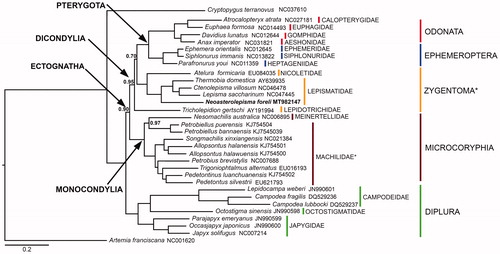
The phylogenetic tree obtained from the Bayesian Inference analysis confirms the results of Leo et al. (Citation2019) and, despite the addition of new sequences, most of the inter-order relationships remain unvaried. A comparison with Chen et al. (Citation2019) and Bai et al. (Citation2020) is, on the other hand, more difficult due to a different taxon sampling in these latter. Overall, the tree supports the monophyly of Ectognatha, Dicondylia, Pterygota, Ephemeroptera, Odonata, Microcoryphia and Diplura with high posterior probability values, but not of Zygentoma (). In fact, as shown in previous morphological and molecular studies (Leo et al. Citation2019; Montagna Citation2020), T. gertschi appears to occupy a basal position within the Dicondylia clade, hence making Zygentoma paraphyletic with respect to Pterygota, although with marginal support. If any, this indicates that the phylogenetic position of this relict species is still a debatable topic (Blanke et al. Citation2014). At the family level, Machilidae (Microcoryphia) is recovered as paraphyletic with respect to Meinertellidae due to the position of Petrobiellus puerensis (nomen nudum) and P. bannaensis (nomen nudum) as sister group to Nesomachilis australica. Within bona fide Zygenoma (i.e. Euzygentoma), Atelura formicaria, the only representative of family Nicoletiidae family, is sister group to the Lepismatidae. Neoasterolepsima foreli appears as the basal group within Lepismatidae at odds with our current understanding of the group that would suggest a closer relationship with Lepisma (Mendes, Citation1991). The study of new samples and the review of previously published ones would be warranted to clarify the position of N. foreli and L. saccharinum.
Figure 1. Phylogenetic tree obtained through a Bayesian statistical approach on the concatenated 13 PCGs of basal Ectognatha and two outgroups (Artemia franciscana and Cryptopygus terranovus). Neoasterolepisma foreli phylogenetic position is bold highlighted. Paraphyletic groups are highlighted with an asterisk. Posterior probabilities are shown at nodes (full support if not otherwise indicated).
Acknowledgments
The authors wish to thank the Dept. of Biotechnology, Chemistry and Pharmacy for providing computational resources and Dr Andrea Bernini for continuous support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MT982147. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA673074 https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP289641 https://www.ncbi.nlm.nih.gov/biosample/SAMN16587686.
Additional information
Funding
References
- Bai Y, Chen J, Li G, Wang H, Luo J, Li C. 2020. Complete mitochondrial genome of the common silverfish Lepisma saccharina (Insecta: Zygentoma: Lepismatidae). Mitochondrial DNA B. 5(2):1552–1553.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Blanke A, Koch M, Wipfler B, Wilde F, Misof B. 2014. Head morphology of Tricholepidion gertschi indicates monophyletic Zygentoma. Front Zool. 11(1):16.
- Boudreaux HB. 1979. Arthropod phylogeny with special reference to insects. New York: Wiley.
- Boore JL, Lavrov DV, Brown WM. 1998. Gene translocation links insects and crustaceans. Nature. 392(6677):667–668.
- Carapelli A, Nardi F, Dallai R, Frati F. 2006. A review of molecular data for the phylogeny of basal hexapods. Pedobiologia. 50(2):191–204.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Chen B, Dong F, Fang W, Xu J, Gu Y, Li M, Sun E. 2019. The complete mitochondrial genome of Ctenolepisma villosa (Insecta: Zygentoma, Lepismatidae). Mitochondrial DNA B. 4(1):1814–1815.
- Comandi S, Carapelli A, Podsiadlowski L, Nardi F, Frati F. 2009. The complete mitochondrial genome of Atelura formicaria (Hexapoda: Zygentoma) and the phylogenetic relationships of basal insects. Gene. 439(1–2):25–34.
- Cook CE, Yue Q, Akam M. 2005. Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc Roy Soc B Biol Sci. 272(1569):1295–1304.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Gaju-Ricart M, Baltanás RM, Bach de Roca C. 2015. Forward without wings: current progress and future perspectives in the study of Microcoryphia and Zygentoma. Soil Org. 87:183–195.
- Kristensen NP. 1998. The groundplan and basal diversification of the hexapods. In: Fortey RA, Thomas RH, editors. Arthropod relationships, systematic association special Volume Series 55. London: Chapman & Hall; p. 281–293.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol E. 34:772–773.
- Leo C, Nardi F, Frati F, Fanciulli PP, Cucini C, Vitale M, Brunetti C, Carapelli A. 2019. The mitogenome of the jumping bristletail Trigoniophthalmus alternatus (Insecta, Microcoryphia) and the phylogeny of insect early-divergent lineages. Mitochondrial DNA B. 4(2):2855–2856.
- Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31(10):1674–1676.
- Mendes LF. 1991. On the phylogeny of the genera of Lepismatidae (Insecta: Zygentoma). Advances in management and conservation of soil fauna. Delhi, Bombay, Calcutta: Oxford & IBH Publications; p. 1–13.
- Mendes LF. 2018. Biodiversity of the Thysanurans (Microcoryphia and Zygentoma). Insect Biodivers: Sci Soc. 2:155–198.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA. p. 1–8.
- Molero-Baltanás R, Bach de Roca C, Tinaut A, Pérez JD, Gaju-Ricart M. 2017. Symbiotic relationships between silverfish (Zygentoma: Lepismatidae, Nicoletiidae) and ants (Hymenoptera: Formicidae) in the Western Palaearctic. A quantitative analysis of data from Spain. Myrmecol News. 24:107–122.
- Montagna M. 2020. Comment on phylogenetic analyses with four new Cretaceous bristletails reveal inter‐relationships of Archaeognatha and Gondwana origin of Meinertellidae. Cladistics. 36(2):227–231.
- Nardi F, Cucini C, Leo C, Frati F, Fanciulli PP, Carapelli A. 2020. The complete mitochondrial genome of the springtail Allacma fusca, the internal phylogenetic relationships and gene order of Symphypleona. Mitochondrial DNA Part B. 5(3):3121–3123.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
