Abstract
In this study, we report the complete mitochondrial genome of Anoplistes halodendri, which covers a total of 15,697 bp in length with 28.27% GC content. The complete mitochondrial genome is composed of 12 protein-coding genes (PCGs) and also contains 22 transfer RNA genes (tRNAs) and two ribosomal RNA genes (rRNAs). Phylogenetic analysis of the A. halodendri with other 21 different species of Cerambycidae indicated that A. halodendri formed an isolated clade and belong to Cerambycinae. The results will be helpful to study the evolutionary relationship among the subfamilies of Cerambycidae.
Anoplistes halodendri is a species of Cerambycidae belong to Coleoptera, which is widely distributed in China, Korea, and Mongolia (Karpiński et al. Citation2018). The host plants of A. halodendri include Tetraena mongolica, Lycium chinense, Hippophae rhamnoides, Lonicera japonica, Pyrus spp., etc. (Wei et al. Citation2012; Karpiński et al. Citation2018). However, the study of A. halodendri on the level of genetic evolution was limited (Wang et al. Citation2019). In this study, we determined the complete mitochondrial genome of A. halodendri and comprehend the evolving relationship by phylogenetic analysis of A. halodendri.
The A. halodendri were collected from Lianjiang, Fujian Province, China (119° 38′ 25″E, 26° 9′ 21″N) and the specimens were stored in the Fujian Agriculture and Forestry University (TN-202,005). The total DNA extraction and purification from A. halodendri with TruSeq DNA Sample Preparation kit (Vanzyme, Nanjing, China) and QIAquick Gel Extraction kit (Qiagen GmbH, Hilden, Germany), respectively. To obtain high-quality short-read data, the purified DNA was quantified by Qubit (Thermo Fisher Scientific Inc., Waltham, MA) and the library was prepared with fragment of 300 bp randomly interrupted. The constructed library was sequenced with 150 bp pair-end reads on the Illumina Hiseq2500 platform according to the manufacturer’s protocol (Illumina, San Diego, CA). After quality control and filtering with Fastp software (Chen et al. Citation2018) of the Illumina data, a total of 53,248,050 clean reads were generated. MtioZ and metaSPAdes software (Nurk et al. Citation2017) were used to assembly the clean reads after quality control and then the assembly sequence was annotated by the MITOS web server (Matthias et al. Citation2013) based on the reference genome of Nortia carinicollis (GenBank accession no. MK863508). The complete mitochondrial genome of A. halodendri was a circular structurally covering a total of 15,697 bp in length (GenBank accession no. MT809475), the overall base composition was 39.6% A, 11.17% G, 17.1% C, and 32.2% T. And the GC content was 28.27%, as well as contains 36 unique genes, including 12 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs).
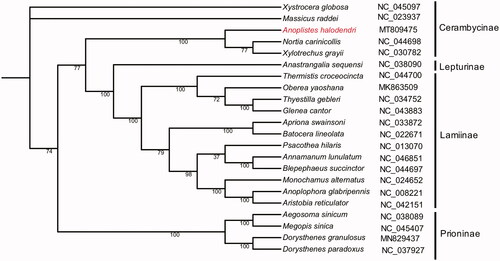
To further explore the phylogenetic position of A. halodendri, a phylogenetic comparison was performed between the A. halodendri and other 21 different species of Cerambycidae. Multiple sequence alignment by the MUSCLE software (Edgar Citation2004), and the phylogenetic trees were inferred using IQ-TREE software (Nguyen et al. Citation2015) with the parameter of -bb 1000. The maximum-likelihood tree showed that A. halodendri formed an isolated clade and belong to Cerambycinae (). The complete mitochondrial genomic data of A. halodendri will be helpful to provide favorable genetic information for realizing the evolutionary relationship between Cerambycidae and other Coleoptera insects.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT809475.
Additional information
Funding
References
- Chen SF, Zhou YQ, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Karpiński L, Szczepański WT, Plewa R, Walczak M, Hilszczański J, Kruszelnicki L, Łoś K, Jaworski T, Bidas M, Tarwacki G. 2018. New data on the distribution, biology and ecology of the longhorn beetles from the area of South and East Kazakhstan (Coleoptera, Cerambycidae). Zookeys. 805(4):59–126.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Peter FS. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Nguyen LT, Schmidt H, von Haeseler A, Minh B. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Wang J, Dai XY, Xu XD, Zhang ZY, Yu DN, Storey KB, Zhang JY. 2019. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ. 7(2):e7633.
- Wei D, Qin Q, Xun F, Liu Q. 2012. EAG and behavioral response of different attractants and field trapping effect on Asias halodendri (Pallas). J Tian Normal Univ. 32(3):71–74.