Abstract
The species Oberonioides microtatantha, belonging to the family Orchidaceae, is a small lithophytic herb endemic in south China with significant conservation values. The complete plastid genome sequence of O. microtatantha reported here is 144,989 bp in length, with a large single copy (LSC) region of 83,920 bp, a small single copy (SSC) region of 13,063 bp, and a pair of inverted repeat (IRa and IRb) regions of 24,003 bp each. The plastome consists of 95 genes, including 72 protein-coding genes, 4 ribosomal RNA genes, and 19 transfer RNA genes. The overall GC content is 36.81%. Phylogenetic analysis placed Oberonioides closet to the genus Liparis in Orchidaceae.
Oberonioides Szlach belongs to the tribe Malaxideae, subfamily Epidendroideae, family Orchidaceae (Chase et al. Citation2015). It comprises of only two species, among which the species O. microtatantha (Schltr.) Szlach. is a small lithophytic herb endemic in China (Szlachetko Citation1995; Chen and Wood Citation2009). It is mainly distributed in southeastern China and is found usually growing on humid and shaded rocks in forests (Chen and Wood Citation2009). Due to its limited distribution, human collection, loss of habitat, and habitat fragmentation, this orchid species was considered near threatened (NT) according to the IUCN Red List of higher plants in China (http://www.iplant.cn/rep/protlist/4).
Samples of O. microtatantha were collected from Guangdong Xiangtoushan Natural Reserve, Guangdong province (China: N23°16′9″, E114°22′26″). Voucher specimen (LYL 305) was deposited in Herbarium of South China Agricultural University (CANT). Total genomic DNA of O. microtatantha was extracted from a mature leaf of an individual by using modified CTAB method (Doyle and Doyle Citation1987). The DNA extracted was directly sequenced using the Illumina HiSeq 2500 Sequencing System. The average read length was 150 bp and the reads were assembled with the software GetOrganelle (Jin et al. Citation2018). Then the genome obtained was annotated by using online software Geseq (Tillich et al. Citation2017). The accession number MT559316 was obtained after submitting the annotated plastid genome of O. microtatantha to the GenBank.
The whole plastid genome of O. microtatantha was 144,989 bp in length, composed by a large single-copy (LSC) region of 83,920 bp, a small single-copy (SSC) region of 13,063 bp, and a pair of inverted repeat (IRa and IRb) regions of 24,003 bp each. The annotated genome comprised 95 genes, including 72 protein-coding genes, 4 ribosomal RNA genes (rrn23, rrn16, rrn5, rrn4.5), and 19 transfer RNA genes. 17 genes were duplicated in the IR regions, including 5 protein-coding genes (rpl2, rpl23, rps19, rps7, ycf2), 4 ribosomal RNA genes (rrn16, rrn23, rrn4.5, rrn5), and 8 transfer RNA genes (trnH-GUG, trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC). The overall GC content of O. microtatantha plastid genome was 36.81% (LSC, 34.1%; SSC, 29.31%; IRs, 43.59%).
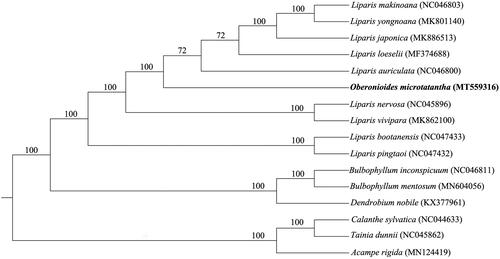
To investigate the phylogenetic location of O. microtatantha within the tribe of Malaxideae, a phylogenetic tree was constructed. The chloroplast genome of O. microtatantha was aligned with 15 other complete chloroplast genomes of Malaxideae, and Acampe rigida, Tainia dunnii, and Calanthe sylvatica of Orchidaceae family as outgroup, using MAFFT ver. 7 (Katoh and Standley Citation2013). Phylogenetic analysis was conducted based on maximum likelihood (ML) analyses using RAxML (Stamatakis Citation2014). Support for the inferred ML tree was inferred by bootstrapping with 1000 replicates and with GTRGAMMA as the nucleotide substitution model. Phylogenetic analysis results suggests that Oberonioides is closely related to the genus Liparis (). The chloroplast genome of O. microtatantha will provide useful genetic information for further study on genetic diversity and conservation of Orchidaceae species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT559316.
Additional information
Funding
References
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Berg CVD, Schuiteman A. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174.
- Chen SC, Wood JJ. 2009. Oberonioides Szlachetko. In: Wu ZY, Raven P, Hong DY, editors. Flora of China 25. St. Louis: Missouri Botanical Garden Press.
- Doyle JJ, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv.256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Szlachetko DL. 1995. Fragmenta Floristicaet Geobotanica. Supplementum. 3:135.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.