Abstract
Completed chloroplast genome of Campanula takesimana Nakai isolated from Dokdo island in Korea is 169,719 bp long (GC ratio is 38.8%) and has four subregions: 102,381 bp of large single-copy (37.8%) and 7,750 bp of small single-copy (32.6%) regions are separated by 29,794 bp of inverted repeat (41.3%) regions including 131 genes (87 protein-coding genes, eight rRNAs, and 36 tRNAs). Phylogenetic analyses suggested that C. takesimana from Dokdo Island form a clade with C. takesimana from Ulleungdo Island and that chloroplast genomes of the two accessions are diverged.
Dokdo Island, located about 216 km off the eastern coast of the Korean peninsula, is a small volcanic island (187,554 m2) with two main islets. The flora of the island with 48 vascular plants is a part of the flora of Ulleungdo Island, a neighboring volcanic island located 87 km northeast from Dokdo Island (Sun et al. Citation2014). Campanula takesimana Nakai (Campanulaceae) is one of two endemic species found on both islands. Chloroplast genomes of C. takesimana from Ulleungdo Island (Cheon et al. Citation2016) and C. punctata, a progenitor species (Yoo et al. Citation2016) were completed. We determined complete chloroplast genome of C. takesimana from Dokdo Island to examine the level of molecular variation of the insular species.
Total DNA of C. takesimana isolated from Dokdo Island, Republic of Korea, was extracted from fresh leaves by using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Voucher was deposited at National Institute of Biological Resources (NIBR) Herbarium (KB) with the accession number, NIBRVP0000752857 (37.24193 N, 131.86464E). Genome was sequenced using NovaSeq6000 at DNALink Inc., Korea, and de novo assembly was conducted by Velvet v1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser v1.12 (Zhao et al. Citation2011), respectively, and using BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009), confirmation of each assembled bases and correctly assembled sequences done together with the primary assembled chloroplast genome based on raw reads generated by Pac-Bio. All bioinformatic analyses were conducted under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr; Park et al., in preparation). Geneious R11 v11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on C. takesimana chloroplast genome (NC_026203).
Chloroplast genome of C. takesimana (GenBank accession is MW013763) from Dokdo Island is 169,719 bp (GC ratio: 38.8%) and has four subregions: 102,381 bp of large single-copy (LSC; 37.8%) and 7750 bp of small single-copy (SSC; 32.6%) regions are separated by 29,794 bp of inverted repeat (IR; 41.3%). It contains 131 genes (87 protein-coding genes (PCGs), eight rRNAs, and 36 tRNAs) and seven pseudogenes (clpP, infA, ycf3, psbJ, and rpl23); 22 genes (nine protein-coding gene, four rRNAs, and seven tRNAs) and two pseudogenes (clpP and infA) are duplicated in IR regions.
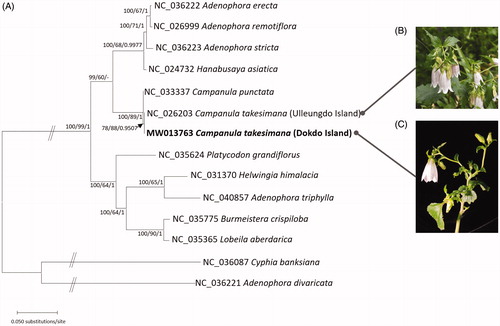
Sixty-five conserved genes extracted from 17 chloroplast genomes of Campanulaceae including two C. takesimana were aligned using MAFFT v7.450 (Katoh and Standley Citation2013). Maximum-likelihood and neighbor-joining trees were reconstructed using MEGA X (Kumar et al. Citation2018) and Bayesian posterior probability of trees was inferred using MrBayes v3.2.7a (Huelsenbeck and Ronquist Citation2001). Phylogenetic trees show that both accessions of C. takesimana are clustered together being sister to C. punctata ().
Figure 1. (A) Maximum-likelihood (bootstrap repeat is 1000) phylogenetic tree of 17 chloroplast genomes of Campanulaceae including two outgroups: Campanula takesimana (MW013763 in this study and NC_026203; Cheon et al. Citation2016), Campanula punctata (NC_033337; Yoo et al. Citation2016), Hanabusaya asiatica (NC_024732; Cheon and Yoo Citation2016), Adenophora stricta (NC_036223; Cheon et al. Citation2017), Adenophora erecta (NC_036222; Cheon et al. Citation2017), Adenophora remotiflora (NC_026999; Kim et al. Citation2016), Adenophora divaricata (NC_036221; Cheon et al. Citation2017), Adenophora triphylla (NC_040857), Platycodon grandifloras (NC_035624; Lin et al. Citation2019), Helwingia himalacia (NC_031370; Yao et al. Citation2016), Burmeistera crispiloba (NC_035775), Lobeila aberdarica (NC_035365), Cyphia banksiana (NC_036087). Neighbor-joining (bootstrap repeat is 10,000) and produced the same topology as the ML tree. The numbers above branches indicate bootstrap support values of maximum-likelihood, neighbor-joining, and Bayesian posterior probability, respectively. (B) Photograph of Campanula takesimana on Ulleungdo Island (taken by Yoonhyuk Bae). (C) Photograph of Campanula takesimana on Dokdo Island (taken by Hwa-Jung Suh).
Compared with the chloroplast genome of C. punctata, C. takesimana from Dokdo Island differed by 33 single nucleotide polymorphisms (SNPs) and 39 insertions and deletions (INDELs) covering 662 bp. Comparison of two C. takesimana chloroplast showed that there were 43 SNPs and 57 INDELs: 28 SNPs/20 INDELs and 2 SNPs/5 INDELs were found in LSC and SSC, respectively. In addition, 13 SNPs/32 INDELs were also identified in the IR regions. There were nine non-synonymous SNPs in four PCGs, ndhK, ndhB, ycf1, and ndhD, two synonymous SNPs in psaA and rpoB, one SNP between two 3-bp INDELs and 8 INDELs of which total length is 126 bp in ycf1 and ycf2. The levels of variation between two accessions of C. takesimana are considered to be high, given the species are restricted on small volcanic islands with small population size.
Plants of C. takesimana on Dokdo Island can be morphologically distinguished from those on Ulleungdo Island by having densely pubescent leaves and funnelform corolla (). Plants of C. takesimana on Dokdo Island bloom in July, lagging one month compared with those on Ulleungdo Island. These differences suggest that the population of C. takesimana on Dokdo Island is diverged from those on Ulleungdo Island. Further detailed morphological and phylogeographic studies are underway to evaluate taxonomic status and understand the origin of C. takesimana on Dokdo Island.
Acknowledgements
We thank to NICEM at Seoul National University in Korea to provide the primary version of raw data of chloroplast genome assembly and annotation of this species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MW013763 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA668862, SAMN16425341, and SRR12816398, respectively.
Additional information
Funding
References
- Cheon K-S, Kim K-A, Jang S-K, Yoo K-O. 2016. Complete chloroplast genome sequence of Campanula takesimana (Campanulaceae), an endemic to Korea. Mitochondrial DNA Part A. 27(3):2169–2171.
- Cheon K-S, Kim K-A, Yoo K-O. 2017. The complete chloroplast genome sequences of three Adenophora species and comparative analysis with Campanuloid species (Campanulaceae). PLOS One. 12(8):e0183652.
- Cheon K-S, Yoo K-O. 2016. Complete chloroplast genome sequence of Hanabusaya asiatica (Campanulaceae), an endemic genus to Korea. Mitochondrial DNA Part A. 27(3):1629–1631.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Kim K-A, Cheon K-S, Jang S-K, Yoo K-O. 2016. Complete chloroplast genome sequence of Adenophora remotiflora (Campanulaceae). Mitochondrial DNA Part A. 27(4):2963–2964.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Lin Y, Jiang S, Yang X. 2019. Characterization of the complete chloroplast genome of Platycodon grandifloras (Campanulaceae: Platycodon), the herbal medicine in China. Mitochondrial DNA Part B. 4(1):2050–2052.
- Sun B-Y, Shin H, Hyun J-O, Kim Y-D, Oh S-H. 2014. Vascular plants of Dokdo and Ulleungdo islands in Korea. Incheon, Korea: National Institute of Biological Resources.
- Yao X, Liu Y-Y, Tan Y-H, Song Y, Corlett RT. 2016. The complete chloroplast genome sequence of Helwingia himalaica (Helwingiaceae, Aquifoliales) and a chloroplast phylogenomic analysis of the Campanulidae. PeerJ. 4:e2734.
- Yoo K-O, Cheon K-S, Kim K-A. 2016. Complete chloroplast genome sequence of Campanula punctata Lam. (Campanulaceae). Mitochondrial DNA Part B. 1(1):184–185.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
