Abstract
In this paper, we determined and characterized the complete mitochondrial genome of Pronghorn spiny lobster Panulirus penicillatus for the first time from South China Sea. The P. penicillatus mitogenome is 15,671 bp long, and consists of 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes (PCGs), and 1 control region. The nucleotide composition of P. penicillatus mitogenome is significantly biased (A, G, T, and C was 33.62, 13.32, 32.31, and 20.75%, respectively) with A + T contents of 65.93%. Almost PCGs used a standard initiation codon or stop codon, except COX2, ND3, ND4 and ND1 were terminated with an incomplete stop codon T and ND5 ended with TA. One microsatellite (C)12 was identified in the control region of P. penicillatus mitogenome sequences. Phylogenetic tree showed that P. penicillatus was first clustered with P. polyphagus and P. versicolor.
Panulirus penicillatus, commonly known as the pronghorn spiny lobster or double-spined rock lobster, is an important commercial species belonging to the family Palinuridae. It is probably the widest global distribution of any species of spiny lobster which known to occur in tropical and sub-tropical areas of the Indo-Pacific region from East Africa and the Red Sea across to Pacific Mexico and Central America (Holthuis Citation1991; Vaitheeswaran Citation2018). Panulirus penicillatus is not gregarious and is nocturnal, usually found in shallow watersat a depth range of 1–4 m in surf-zones of coral reefs and large rocky outcroppings. It is heavily exploited for food throughout its wide range, this does not seem to have had a large impact on total populations. Wide-ranging stock assessments and biological reference points such as population ecology, growth, and minimum suitable catch size, sexual maturity and reproduction season have been examined to support a modest fishery of P. penicillatus (Ebert and Ford Citation1986; Plaut Citation1993; Hogarth and Barratt Citation1996; Chauvet and Coutures Citation2001; Szuwalski et al. Citation2016). Early studies mainly focused on the developmental and reproductive biology of P. penicillatus. Recently, the researchers have paid more attention to its genetics including the genetic isolation between the western and eastern Pacific populations, genetic diversity and population structure, range-wide phytogeography and development of compound microsatellite markers (Chow et al. Citation2011; Abdullah et al. Citation2014, Citation2017; Iacchei et al. Citation2016). Morphological differences among phyllosoma larvae from the different Pacific populations also showed that there are significant geographic differences same as the above genetic analysis (Matsuda et al. Citation2019a, Citation2019b).
The specimens of P. penicillatus were collected from Qionghai, China (N19°18′48.37″, E110°40′20.21″), and stored in the marine crustacean specimen room (C20191119PP) in Qionghai research base of Hainan Academy of Ocean and Fisheries Sciences. The libraries with an average length of 350 bp were constructed using the NexteraXT DNA Library Preparation Kit, and sequencing was performed on the Illumina Novaseq platform (Total Genomics Solution Limited, SZHT) with the 150 bp average length of the generated reads. The complete mitochondrial genome of P. longipes were assembled with 5.91 G clean reads using the de novo assembler SPAdes 3.11.0, and annotated using the MITOS (http://mitos.bioinf.uni-leipzig.de/index.py). Based on nucleotide sequences of 20 Achelata species mitogenome available in the GenBank (Table S2), a phylogenetic analysis was carried out using IQ-TREE(Nguyen et al. Citation2015) v1.6.12 to investigate the evolution position of P. penicillatus using the maximum–likelihood (ML) method with 1000 bootstrap replicates.
The whole mitogenome of P. penicillatus (Table S1) is 15,671 bp in size (GenBank Accession No. MT533488). The base content was 32.80% A, 12.01% G, 32.63% T, and 22.56% C. The 65.43% of (A + T) showed great preference to AT. It consists of 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes (PCGs), and 1 control region. Four PCGs (ND1, ND4, ND4L, and ND5), eight tRNA genes and two rRNA genes were located on the light strand, the others were encoded by the heavy strand.
The 22 tRNA genes in P. penicillatus mitogenome vary in length from 63 to 72 bp. tRNA-Leu and tRNA-Ser both have two type copies respectively. The 12S rRNA is 863 bp and located between tRNA-Val and the control region, and the 16S rRNA is 1291 bp, located between tRNA-Val and tRNA-Leu. All 13 PCGs use a normal initiation codon ATN or TTG. Simultaneously, eight PCGs were terminated with a usual stop codon in addition five PCGs using a normal stop codon (COX2, ND3, ND4 and ND1 use a single T; ND5 use TA). The control region is 731 bp, located between 12S rRNA and tRNA-Ile. Interestingly, we identified one microsatellite (SSR) in P. penicillatus mitogenome using MISA (Beier et al. Citation2017), a (C)12 is located in the control region which was also found similar in some closely related species with different number of repetitions ().
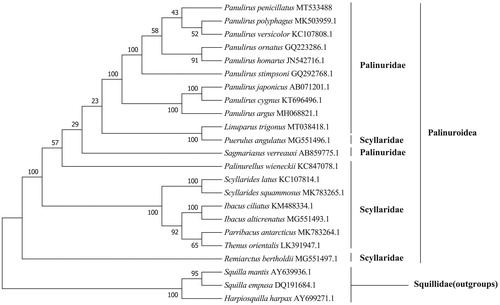
The phylogenetic tree () showed that P. penicillatus was formed a clade by first clustering with P. polyphagus and P. versicolor, and further clarified the phylogenetic relationships of the species of lobsters in the family Palinuridae.
Harpiosquilla harpax, Squilla empusa, and Squilla mantis were used as outgroups.
Supplemental Material
Download Zip (8.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in “figshare” at https://doi.org/10.6084/m9.figshare.12812081.v1.
Additional information
Funding
References
- Abdullah MF, Cheng J-H, Chen T-I, Imai H. 2017. Development of compound polymorphic microsatellite markers for the pronghorn spiny lobster Panulirus penicillatus and comparison of microsatellite data with those of a previous mitochondrial DNA study performed in the northwestern Pacific. Biogeography. 19:61–68.
- Abdullah MF, Chow S, Sakai M, Cheng J-H, Imai H. 2014. Genetic diversity and population structure of Pronghorn Spiny Lobster Panulirus penicillatus in the Pacific Region1. Pacific Sci. 68(2):197–211.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Chauvet C, Coutures E. 2001. Growth and minimum suitable catch size of spiny lobsters, Panulirus penicillatus (Olivier, 1791) and Panulirus longipes bispinosus Borradaile, 1899 (Decapoda, Palinuridae) in the southern lagoon of New Caledonia. Crustac. 74(11):1189–1199.
- Chow S, Jeffs A, Miyake Y, Konishi K, Okazaki M, Suzuki N, Abdullah MF, Imai H, Wakabayasi T, Sakai M. 2011. Genetic isolation between the western and eastern Pacific populations of pronghorn spiny lobster Panulirus penicillatus. PLoS One. 6(12):e29280.
- Ebert TA, Ford RF. 1986. Population ecology and fishery potential of the spiny lobster Panulirus penicillatus at Enewetak Atoll, Marshall Islands. Bull Mar Sci. 38:56–67.
- Hogarth P, Barratt L. 1996. Size distribution, maturity and fecundity of the spiny lobster Panulirus penicillatus (Olivier 1791) in the Red Sea. Tropical Zool. 9(2):399–408.
- Holthuis LB. 1991. Marine lobsters of the world. An annotated and illustrated catalogue of species of interest to fisheries known to date. FAO fisheries Synopsis No. 125. 13:292.
- Iacchei M, Gaither MR, Bowen BW, Toonen RJ. 2016. Testing dispersal limits in the sea: range-wide phylogeography of the pronghorn spiny lobster Panulirus penicillatus. J Biogeogr. 43(5):1032–1044.
- Matsuda H, Sakai M, Yanagimoto T, Chow S. 2019a. Morphological differences among phyllosoma larvae of the pronghorn spiny lobster, Panulirus penicillatus, from the western, central, and eastern areas of the Pacific Ocean. Crust Res. 48(0):105–118.
- Matsuda H, Sakai M, Yanagimoto T, Chow S. 2019b. Morphological differences between phyllosoma larvae from the western–central and eastern Pacific populations of the pronghorn spiny lobster Panulirus penicillatus. bioRxiv. :667469.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Plaut I. 1993. Sexual maturity, reproductive season and fecundity of the spiny lobster Panulirus penicillatus from the Gulf of Eilat (Aqaba), Red Sea. Mar Freshwater Res. 44(4):527–535.
- Szuwalski CS, Castrejon M, Ovando D, Chasco B. 2016. An integrated stock assessment for red spiny lobster (Panulirus penicillatus) from the Galapagos Marine Reserve. Fish Res. 177:82–94.
- Vaitheeswaran T. 2018. On rare occurrence of pronghorn spiny lobster, Panulirus penicillatus (Olivier, 1791) off Tuticorin coast, India (08 35.912’N and 78 25.327’E)(25 M). J Agric Sci Bot. 02(01):7–8.