Abstract
Vitis vinifera × Vitis labrusca ‘Shenhua’ is a tetraploid grape, a Franco-american species. This study first published the complete chloroplast genome of Vitis vinifera × Vitis labrusca ‘Shenhua’ was assembled. The chloroplast genome is 160928 bp in length, including a large single copy region (89,148 bp), a small single-copy region (19,072 bp) and a pair of inverted repeats of 26,354 bp. The chloroplast genome encodes 133 genes, comprising 88 CDSs, 37 tRNA genes and 8 rRNA genes. The phylogenetic tree demonstrated that Vitis vinifera × Vitis labrusca ‘Shenhua’ is different from the other 16 varieties.
Vitis vinifera × Vitis labrusca ‘Shenhua’ shows strong heat-resistant and disease resistance. Thus, it is widely used for breeding of heat-resistant grape varieties. It is also an excellent wine and food grape variety because of its high anthocyanin, titratable acids and soluble solids contents (Zha et al. Citation2020). The complete chloroplast genome of Vitis vinifera × Vitis labrusca ‘Shenhua’ was assembled (GenBank: MT916288) and can be perform to phylogenetic analysis. It supplies significant information for the basic research of grape resistance breeding.
Genomic DNA was extracted from leaves of Vitis vinifera × Vitis labrusca ‘Shenhua’ originated from the Zhuang-hang Experimental Station of Shanghai Academy of Agricultural Sciences (30°89’N, 121°39’W) and stored at the Center for Viticulture and Enology, Shanghai Jiao Tong University, the sample was called ‘Shenhua’. This DNA was used to prepare 400 bp tiny fragment DNA library and then sequenced by the NovaSeq-PE150 sequencing platform (Illumina, CA, USA). 3.49 Gb clean reads were obtained in the aggregate. The complete chloroplast genome was composed by SPAdesv3.9.0 (Bankevich et al. 2012) and A5-MiSeqv20150522 (Coil et al. Citation2015) software. Genome functional annotation is done on the website (https://chlorobox.mpimp-golm.mpg.de/geseq.html). The complete chloroplast genome of Vitis vinifera (GenBank; DQ424856) serve as a reference (Jansen et al. Citation2006).
The length of chloroplast genome is 160,928 bp, consistsing of a large single copy region (89,148 bp), a small single-copy region 19,072 bp and a pair of inverted repeats of 26,354 bp. The chloroplast genome includes 133 single genes, including 88 protein-coding genes (CDS), 37 tRNA and 8 rRNA genes. 37.38% and 62.62% are the amounts of GC content and AT content of the grape genome. In these genes, most of them are single-copy, whereas 7 CDS (rp123, rp12, rps7, rps12, ndhB, ycf2, ycf15), 4 rRNAs (rrn4.5, rrn-16, rrn5, rrn-23) and 7 tRNAs (trnL-CAA, trnI-CAU, trnV-GAC, trnA-UGC, trnI-GAU, trnR-ACG, trnN-GUU) appear as double-copies.
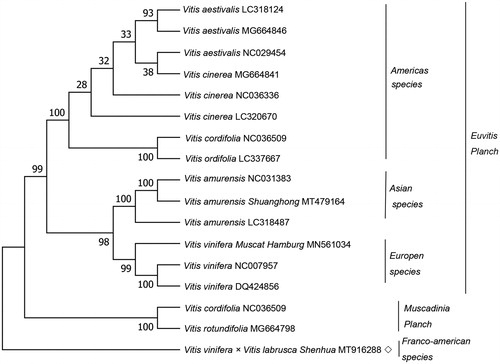
A Maximum Likelihood (ML) phylogenetic tree was builded by using 16 Vitis species through the MEGA7 (Kumar et al. Citation2016) to affirm the phylogenetic position of Vitis vinifera × Vitis labrusca ‘Shenhua’ within the family Vitaceae. The phylogenetic tree demonstrated that the 17 Vitis species are divided into two classes (), which concordant with the former report (Xie et al. Citation2017). The phylogenetic tree demonstrated that Vitis vinifera × Vitis labrusca ‘Shenhua’ is different from the other 16 varieties.
Supplemental Material
Download TIFF Image (12 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank: MT916288 at https://www.ncbi.nlm.nih.gov/genbank/.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich A A, Dvorkin M, Kulikov A S, Lesin V M, Nikolenko S I, Pham S, Prjibelski A D, et al. 2012. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 19(5):455–477.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Jansen RK, Kaittanis C, Saski C, Lee S-B, Tomkins J, Alverson AJ, Daniell H. 2006. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 6(1):32.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Xie H, Jiao J, Fan X, Zhang Y, Jiang J, Sun H, Liu C. 2017. The complete chloroplast genome sequence of Chinese wild grape Vitis amurensis (Vitaceae: Vitis L.). Conservation Genet Resour. 9(1):43–46.
- Zha Q, Xi XJ, He YN, Jiang AL. 2020. Transcriptomic analysis of the leaves of two grapevine cultivars under high-temperature stress. Sci Hortic. :265.109265.