Abstract
Camellia rhytidophylla is an endangered plant with economic value. Using Illumina sequencing, the chloroplast genome of C. rhytidophylla was sequenced and analyzed in this study. The complete chloroplast genome is 157,073 bp in length, which consisted of a pair of inverted repeat regions of 26,055 bp (IRa and IRb) separated by a large single-copy region (LSC) of 86,680 bp and a small single-copy region (SSC) of 18,283 bp. The C. rhytidophylla chloroplast genome encodes 135 genes, including 87 protein-coding genes, 37 tRNA genes, 8 rRNA genes, and 3 pseudogenes. Sequence comparison analysis with the chloroplast sequences of 28 other Camellia plants found that C. rhytidophylla had the closest relationship with C. szechuanensis. This study provides a theoretical basis for the analysis of the distant relationship of Camellia.
Camellia rhytidophylla belongs to the genus Camellia in the family Theaceae. C. rhytidophylla was planted in the Environmental Horticulture Research Institute of the Guangdong Academy of Agricultural Sciences (N23°23′, E113°23′, Guangzhou, China) (No: EHRIGAASC003). C. rhytidophylla can be used for landscaping. Evergreen shrubs with thick, leathery leaves and uneven leaf surface. White flowers, solitary in the axillary parietal lobe. The fruit is nearly spherical with irregular nodular protrusions on the surface.
The chloroplast genome DNA of C. rhytidophylla was extracted from young leaves. Firstly, DNA was broke into fragments of 300 bp length using Covaris M220 (Covaris, Woburn, MA), and then, we constructed shotgun sequencing libraries according to the TruSeq™ DNA Sample Prep Kit for Illumina. Finally, whole genome sequencing was executed using the Illumina NovaSeq platform (Illumina, USA) (Genepioneer Biotechnologies Co. Ltd, Nanjing, China). Pair-end Illumina raw reads were filtered using Trimmomatic (Bolger et al. Citation2014), and these reads were mapped to the chloroplast genome of the reference species (Genbank accession number: NC_024663), using Bowtie2 v2.2.4 (Langmead and Salzberg Citation2012) to exclude reads of nuclear and mitochondrial origins. SPAdes 3.6.1(Bankevich et al. Citation2012) and Sequencher 5.3.2 (Gene Codes Inc., Ann Arbor, MI, USA) were used for de novo assembly to reconstruct the chloroplast genomes. A ‘genome walking’ technique was used to remove gaps (Souza et al. Citation2019). Jellyfish v.2.2.3 (Marcais and Kingsford Citation2011) was used to correct misassembled contigs. CpGAVAS (Liu et al. Citation2012) was used for annotation of the chloroplast genomes and OGDRAW (Lohse et al. Citation2007) was used to draw a circular representation. The complete chloroplast genome sequence has been submitted to Genbank with the accession number of MT663343.
The complete chloroplast genome sequence of C. rhytidophylla is 157,073 bp in length, containing a LSC (86,680 bp) region, a SSC (18,283 bp) region, and two inverted repeat regions (IRa and IRb, each 26,055 bp). The GC content of the overall chloroplast genome, LSC, SSC, and IR regions are 37.31, 35.32, 30.61, and 42.97%, respectively. The GC content of the two IR regions is higher than those of the SSC and LSC, which is similar to Spathiphyllum ‘Parrish’ (Liu et al. Citation2019a), Spathiphyllum cannifolium (Liu et al. Citation2019b), and Celosia cristata (Liu et al. Citation2020). The chloroplast genome is made up of 135 genes in total, including 87 protein-coding genes, 37 tRNAs, 8 rRNAs, and 3 pseudogenes.
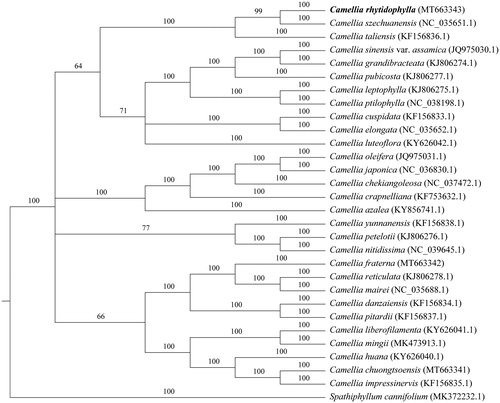
The whole genome was used for phylogenetic tree analysis. First, we use MAFF v7.427 (Katoh et al. Citation2005) auto mode to align each sequence. The gaps in the alignment were removed using the program trimAl with ‘-nogaps’ v 1.4 (Capella-Gutierrez et al. Citation2009). Finally, MrBayes v3.2.7 (Fredrik et al. Citation2012) was used to construct the phylogenetic tree (). And the result suggested that C. rhytidophylla had the closest relationship with C. szechuanensis. This study will be useful for further analysis of genetic diversity, molecular markers, and molecular breeding in Camellia.
Author contributions
Performed the experiments investigation, project administration, writing the original draft and data curation: XL.
Prepared the resources: YS LH YX CZ.
Supervised the project and made revisions to the manuscript: BY.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly obtained at this study are available in the NCBI under accession number of MT663343 (https://www.ncbi.nlm.nih.gov/nuccore/MT663343).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Fredrik R, Maxim T, Paul VDM, Daniel LA, Aaron D, Sebastian H, Bret L, Liang L, Marc AS, John PH. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542.
- Katoh K, Kei-Ichi K, Hiroyuki T, Takashi M. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and Genbank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13:715.
- Liu XF, Ye YJ, Liu JM, Yu B, Xu YC. 2020. Complete chloroplast genome sequence and phylogenetic analysis of Celosia cristata ‘Xiaguang’. Mitochondrial DNA Part B. 5(2):1338–1339.
- Liu XF, Zhu GF, Li DM, Wang XJ. 2019a. Complete chloroplast genome sequence and phylogenetic analysis of Spathiphyllum 'Parrish'. PLOS One. 14(10):e0224038.
- Liu XF, Zhu GF, Li DM, Wang XJ. 2019b. The complete chloroplast genome sequence of Spathiphyllum cannifolium. Mitochondrial DNA Part B. 4 (1):1822–1823.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5-6):267–274.
- Marcais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 27(6):764–770.
- Souza UJBd, Nunes R, Targueta CP, Diniz-Filho JAF, Telles MPC. 2019. The complete chloroplast genome of Stryphnodendron adstringens (Leguminosae – Caesalpinioideae): comparative analysis with related Mimosoid species. Sci Rep. 9(1):14206.