Abstract
Here we generated the complete mitochondrial genome of one subspecies of R. affinis (R. affinis himalayanus) using next generation sequencing and Sanger sequencing. The length of the complete mitochondrial genome was 16,886 bp, containing 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and a non-coding control region. A maximum-likelihood tree based on the 13 concatenated mitochondrial protein-coding genes of 16 Rhinolophus taxon and one outgroup Hipposideros armiger indicates that R. affinis shows a closer relationship with R. sinicus complex than with other taxa.
The intermediate horseshoe bat, Rhinolophus affinis, is distributed throughout Southeast Asia and nine subspecies have been recognized (Csorba et al. Citation2019). There are three subspecies of R. affinis in China, R. a. himalayanus, R. a. macrurus and R. a. hainanus (Mao et al. Citation2010). Previous studies on this species have identified a hybrid zone between R. a. himalayanus and R. a. macrurus in the eastern region of China (Mao et al. Citation2013; Mao and Rossiter Citation2020). In this study we generated the complete mitochondrial genome of R. a. himalayanus (GenBank Accession number: MT845219).
An adult male of R. a. himalayanus was captured with nets from Anhui province, China (30°26′51.82″N, 118°26′54.16″E) and stored in 95% ethanol at −80 °C laboratory freezer in East China Normal University (Voucher No. AH17). Our sampling procedure was approved by the National Animal Research Authority, East China Normal University (approval ID bf20190301). Muscle tissue was used to extract genomic DNA using DNeasy Blood & Tissue Kit (Tiangen, China). Then a DNA library was constructed with insert fragments of ∼350bp and sequenced on the Illumina HiSeq 4000 sequencer (150 bp paired-end). A total of 688,763,636 reads were generated and processed using TRIMMOMATIC-0.36 (Bolger et al. Citation2014) with a sliding window of 4 bp, minimum average PHRED quality score of 20 and minimum reads length of 50 bp. Over six hundred million filtered reads were obtained and mapped to the complete mitochondrion genome of Rhinolophus sinicus sinicus (GenBank accession no. KP257597) using BWA v. 0.7.12-r1039 (Li and Durbin Citation2009) and SAMTOOL v. 1.9 (Li and Durbin Citation2009) was used to retrieve mapped mitochondrial reads of R. affinis. Then the retrieved reads were used to assemble mitochondrial genome by A5-miseq version 20160825 with default parameters (Coil et al. Citation2015). We amplified the non-coding control region (D-loop) using traditional Sanger sequencing. PCR primers have been described previously (Sun et al. Citation2009).
The complete mitochondrial genome of R. affinis is 16,886 bp in length and was annotated based on published mitochondrion genome of Rhinolophus sinicus sinicus. It includes 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and a non-coding control region (D-loop). Gene number and order are identical with other Chiroptera mitogenomes (e.g. Xing and Mao Citation2016; Wang et al. Citation2020). The overall base composition of the whole mitochondrial genome is 30.98% A, 25.13% T, 28.96% C and 14.92% G. All genes were initiated with ATN (nd1, cox1, cox2, atp8, atp6, cox3, nd4, nd6 and cytb with ATG; nd2, nd3 and nd5 with ATA) except for nd4l with GTG. Eight coding genes use TAN as the termination codons (cox1, atp8, atp6, nd3, nd4l, nd5, and nd6 with TAA; cox2 with TAG), and only one gene (cytb) is stopped with AGA. Incomplete stop codon (T–– or TA–) was found in nd1, nd2, cox3, and nd4.
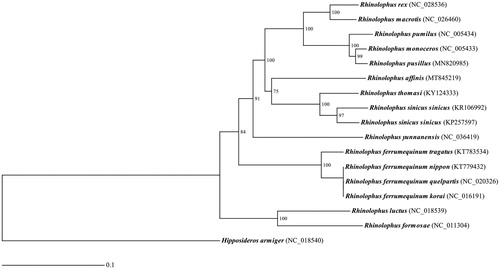
A maximum-likelihood tree was constructed using RAxML v.8.2.11 (Stamatakis Citation2014) based on the 13 concatenated mitochondrial protein-coding genes of 16 Rhinolophus taxa and one outgroup Hipposideros armiger () with GTRGAMMA model and bootstraps 1000 setting. The ML tree indicates that R. affinis shows a closer relationship with R. sinicus complex (R. sinicus and R. thomasi) ().
Figure 1. A maximum-likelihood tree reconstructed based on the 13 concatenated mitochondrial protein-coding genes from 17 bat species. Hipposideros ammiger was used as an outgroup. Numbers at the nodes indicated bootstrap support values. GenBank accession number for each bat mitogenome is indicated in bracket.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, GenBank Accession number: MT845219.
Additional information
Funding
References
- Bolger A, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Csorba G, Hutson AM, Rossiter SJ, Burgin C. 2019. Family Rhinolophidae (Horseshoe Bats). In: Mittermeier RA, Wilson DE, editors. Handbook of the mammals of the World: 9. Chiroptera. Barcelona (Spain): Lynx Ediciones; p. 1–22.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Mao X, He G, Hua P, Jones G, Zhang S, Rossiter SJ. 2013. Historical introgression and the persistence of ghost alleles in the intermediate horseshoe bat (Rhinolophus affinis). Mol Ecol. 22(4):1035–1050.
- Mao X, Rossiter SJ. 2020. Genome-wide data reveal discordant mitonuclear introgression in the intermediate horseshoe bat (Rhinolophus affinis). Mol Phylogenet Evol. 150:106886.
- Mao X, Zhu G, Zhang S, Rossiter SJ. 2010. Pleistocene climatic cycling drives intra-specific diversification in the intermediate horseshoe bat (Rhinolophus affinis) in Southern China . Mol Ecol. 19(13):2754–2769.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun K, Feng J, Jin L, Liu Y, Shi L, Jiang T. 2009. Structure, DNA sequence variation and phylogenetic implications of the mitochondrial control region in horseshoe bats. Mamm Biol. 74(2):130–144.
- Xing Y, Mao X. 2016. The complete mitochondrial genome of the Thomas’s horseshoe bat (Rhinolophus thomasi) using next-generation sequencing and Sanger sequencing. Mitochondrial DNA Part B. 1(1):964–965.
- Wang J, Zhao A, Sun H. 2020. The complete mitochondrial genome of the least horseshoe bat (Rhinolophus pusillus). Mitochondrial DNA Part B. 5(1):881–882.
