Abstract
The complete mitochondrial genome of medicinal plant, Euonymus alatus, was sequenced for the first time. The genome sequence is 1,045,106 bp in length (GenBank accession number MW009108), with 44.98% GC contents. There are 72 genes in the genome, including 41 known protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), and three ribosomal RNAs (rRNAs). The phylogenetic trees of 28 species are constructed using the maximum-likelihood method. The information will provide references for phylogenetic research.
Euonymus alatus is an important medicinal plant of Euonymus L., which is distributed over China, Japan and Korea (Qin et al. Citation2011). Its branches with thrombus wings are the medicinal parts and are commonly called ‘Guijianyu’ in China (National Pharmacopoeia Commission Citation1963). Many chemical constituents have been isolated and identified from E. alatus, including sesquiterpenoids, diterpenoids, triterpenoids, flavonoids, phenylpropanoids, lignans, steroids, alkaloids and other compounds (Zhou et al. Citation2009; Zhai et al. Citation2016). Some studies have shown that the extracts and compounds from E. alatus have a wide pharmacological effects, including anti-diabetes, anti-inflammatory, anti-tumor, liver protection and so on (Fan et al. Citation2020). At present, there are reports on molecular studies of Celastraceae, but the related research of E. alatus is quite limited (Simmons et al. Citation2001).
Fresh leaves of E. alatus were collected from Dandong, China (E 124°39′79.85″, N 40°15′08.55″). Professor Tingguo Kang of Liaoning University of Traditional Chinese Medicine identified the certificate specimen (E. alatus number: 10162200522008LY). The plant samples were deposited in the herbarium of Liaoning University of Traditional Chinese Medicine, and the genomic DNA was stored in the Key Laboratory of Traditional Chinese Medicine. Total genomic DNA was extracted with a Plant Tissue Mitochondrial DNA Extraction Kit (Genmed Scientific Inc., Arlington, MA, USA). One short sequencing library (ca. 300 bp) were constructed and sequenced using Illumina Novaseq 6000 platform. Genome assembly was performed by SOAP denovo 2.04 (Luo et al. Citation2012). The mitochondrial genes were annotated using the online DOGMA tool (http://dogma.ccbb.utexas.edu/index.html) (Wyman et al. Citation2004).
The assembly of the E. alatus resulted in a final sequence of 1,045,106 bp in length (GenBank accession number MW009108), and GC content was 44.98%. The structure of mitochondrial genome was a typical circle type. There were 72 genes annotated, including 41 known protein-coding genes, 22 tRNAs, and three rRNAs. The total length of protein-coding genes was 33,960 bp, accounting for 3.25% of the total genome length. The average length of tRNA was 75 bp, while the rRNA was 1828 bp. In addition, we found that 10 genes (nad2, nad7, nad5, nad1, nad4, cox2, ccmC, ccmFc, rps3, rps10) contained 24 introns.
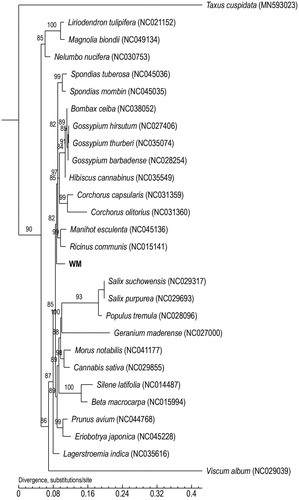
Phylogenetic trees are widely used in genetic and evolutionary studies of various organisms. Using the maximum-likelihood method, we constructed a phylogenetic tree based on the complete mitochondrial genome, including 27 angiosperms (including E. alatus, WM) and one species (Taxus cuspidata) of outgroup. We also listed all species accession numbers used in this study. The maximum-likelihood tree showed the position of E. alatus in all species, and E. alatus can be clustered into a single branch. Additionally, T. cuspidata was far from the other species (). The phylogenetic tree constructed based on the mitochondrial genome sequence of E. alatus will help its classification and evolutionary research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/search/all/?term=MW009108, reference number MW009108.
Additional information
Funding
References
- Fan L, Zhang C, Ai L, Wang L, Li L, Fan W, Li R, He L, Wu C, Huang Y. 2020. Traditional uses, botany, phytochemistry, pharmacology, separation and analysis technologies of Euonymus alatus (Thunb.) Siebold: a comprehensive review. J Ethnopharmacol. 259:112942.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1(1):18
- National Pharmacopoeia Commission. 1963. People's Republic of China Pharmacopoeia. Part I. Beijing: China Medical Science and Technology Press.
- Qin XY, Yan SX, Wei FY. 2011. Geographical distribution of plants of Celastraceae in China. J Northeast Forestry Univ. 39(1):120–123.
- Simmons MP, Savolainen V, Clevinger CC, Archer RH, Davis JI. 2001. Phylogeny of the Celastraceae inferred from 26S nuclear ribosomal DNA, phytochrome B, rbcL, atpB, and morphology. Mol Phylogenet Evol. 19(3):353–366.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhai XF, Lenon GB, Xue CCL, Li CG, Saad B. 2016. Euonymus alatus: a review on its phytochemistry and antidiabetic activity. Evid-Based Compl Alt. 2016:1–12.
- Zhou X, Liu Y, Gong XJ, Zhao C, Chen HG. 2009. Studies on chemical constituents of the Euonymus alatus (Thunb.) Sieb. II. Chin Pharm J. 44(18):1375–1377.