Abstract
Peach (Prunus persica) is one of the most important economic fruit crops in the current society. In this study, we reported the complete chloroplast genome sequence of peach cultivar ‘Rui Guang 18’ (Prunus persica var. nectarina cv. ‘Rui Guang 18’) using whole genome sequencing data. The complete chloroplast genome size was 157,494 bp as a circle, which contained a large single-copy (LSC) region of 85,848 bp, a small single-copy (SSC) region of 18,983 bp, and a pair of inverted repeats (IRs) region of 52,663 bp. The total GC content of the chloroplast genome is 36.8%. The chloroplast genome contains 116 genes, including 76 protein coding genes (PCG), 6 ribosome RNA (rRNA) genes, and 35 transfer RNA (tRNA) genes. The complete chloroplast genome will be a potential genetic resource for peach breeding.
The cultivated peach (Prunus persica), which belongs to the Rosaceae family, is a perennial tree and an economically important fruit crop, and it contains four botanical varieties, namely Prunus persica var. nectarina (Ait.) Maxim., Prunus persica var. compressa (Lond.) Bean, Prunus persica var. densa Makino, and Prunus persica var. duplex Rehd. (Li et al. Citation1997; Yu et al. Citation2018). In China, the planting area of peach is about 860,000 hectares and its annual yield is 15 million tons. And peach is the fourth largest economic fruit, which produces enormous economic value every year. Research has shown that early peaches were selected and domesticated at least 7500 years ago in the lower Yangzi River valley of China (Zheng et al. Citation2014). Nectarine is a mutant of peach, which was cultivated over 2000 years (Yoon et al. Citation2006). The fruit of nectarine is characterized by hairless, delicious flavor, fragrant, sweet and crisp texture, as well as low calorific value and high antioxidant capacity (Jayarajan et al. Citation2019). In China, ‘Rui Guang 18′ is an outstanding medium-mature sweet nectarine, and its fruit with large size, yellow flesh and deep red skin color. Chloroplast is the important organelle of photosynthesis in green plants cells which can convert light energy into the energy-rich adenosine tri-phosphate (ATP) and sugars. As a result, carrying out chloroplast genome research could provide research basis for plant growth and fruit development. In this study, we reported the complete chloroplast genome sequence of peach cultivar ‘Rui Guang 18′ using high-throughput sequencing method. The annotated chloroplast genome sequence was submitted to GenBank under the accession number MW033980.
The fresh and healthy leaves of ‘Rui Guang 18′ were collected in National GermplasmRepository of Peach and Strawberry (Beijing), Beijing Academy of Forestry and Pomology Sciences, Beijing, China (39°96′N, 116°22′E). Genomic DNA was extracted from the fresh leaves according to a modified CTAB method (Doyle and Doyle Citation1987). The library with insert size of 350 bp fragments was constructed. The high throughput sequencing was carried out using third-generation sequencing (TGS) on the OXFORD_NANOPORE PromethION platform. The complete chloroplast genome of Prunus persica (https://www.ncbi.nlm.nih.gov/nuccore/NC_014697.1) was used as the reference sequence to assembled and annotated. We used minimap2 software to compare reference chloroplast genome sequence to the raw data and then used canu software to assemble the selected data and corrected the errors. Nucmer software was used to compare the reference genome with the corrected genome, and mummerplot was used to draw the collinearity map, then nucmer software was used to draw the collinearity graph again. The complete chloroplast genome was annotated with the Geneious (Kearse et al. Citation2012).
Our result showed that the complete chloroplast genome of ‘Rui Guang 18′ was 1,57,494 bp as a circle, which was slightly shorter than the reference chloroplast genome sequence (1,57,790 bp). It contained a large single copy (LSC) region of 85,848 bp, a small single copy (SSC) region of 18,983 bp, and a pair of inverted repeats (IRs) region of 52,663 bp. The total GC content of the chloroplast genome is 36.8%. The chloroplast genome contains 116 genes, including 76 protein coding genes (PCG), 6 ribosome RNA (rRNA) genes, and 35 transfer RNA (tRNA) genes. Among these genes, 17 protein coding genes (atpF, clpP, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU,trnL-UAA, trnV-UAC and ycf3) had introns, and two of these genes (clpP and ycf3) had two introns, whereas the others had a single intron. 2 protein coding genes (rpl23 and rps7), 2 rRNA genes (rrn4.5 and rrn16) and 5 tRNA genes (trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU and trnV-GAC) were duplicated, the others only had a single copy. 6 protein coding genes (ndhB, rpl23, rps7, ycf1, ycf15, and ycf68), 2 rRNA genes (rrn4.5, rrn5, rrn16 and rrn23) and 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC) were located at the IR regions. Especially, the ycf1 was trans-spliced genes, which started in LSC region and ended in IR region.
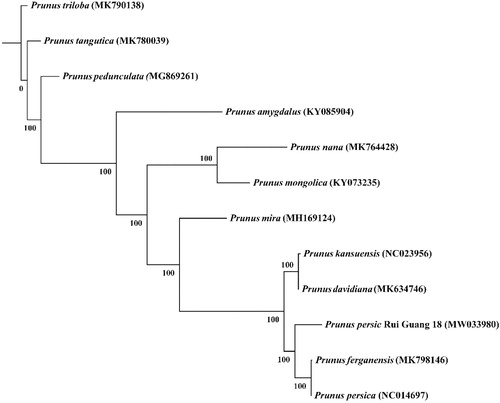
All the complete chloroplast genome of 11 species belonged to the genus Amygdalus L. were used for phylogenetic reconstruction. The complete chloroplast genome sequences were used to construct phylogenic maximum likelihood (ML) trees using RaxML software v 8.2.9 (Stamatakis Citation2014). The bootstrap values of ML trees were calculated using 1000 replicates. The best-fit model was selected using ModelFinder (Kalyaanamoorthy et al. Citation2017). Our result of the phylogenetic analysis () showed that the Prunus persica ‘Rui Guang 18′ is closely related to the Prunus persica and Prunus ferganensis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW033980.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Li SH, Xiao XG, Jia KG. 1997. Cultivation of peaches and nectarines (in Chinese). Beijing, China: Higher Education Press.
- Jayarajan S, Sharma RR, Sethi S, Saha S, Sharma VK, Singh S. 2019. Chemical and nutritional evaluation of major genotypes of nectarine (Prunus persica var nectarina) grown in North-Western Himalayas. J Food Sci Technol. 56(9):4266–4273.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yoon J, Liu DC, Song W, Liu WS, Zhang AM, Li SH. 2006. Genetic diversity and ecogeographical phylogenetic relationships among peach and nectarine cultivars based on simple sequence repeat (SSR) markers. JASHS. 131(4):513–521.
- Yu Y, Fu J, Xu YG, Zhang JW, Ren F, Zhao HW, Tian SL, Guo W, Tu XL, Zhao J, et al. 2018. Genome re-sequencing reveals the evolutionary history of peach fruit edibility. Nat Commun. 9(1):1–13.
- Zheng Y, Crawford GW, Chen X. 2014. Archaeological evidence for peach (Prunus persica) cultivation and domestication in China. PLOS One. 9(9):e106595.