Abstract
The complete chloroplast genome of a local rare fern species Drynaria acuminata was sequenced. The genome has a length of 151,591 bp with 40.8% GC content, with in total 131 genes were annotated, including 88 protein genes, 35 tRNAs, and 8 rRNAs. This work provides basic information for its phylogeographical and conservation research.
Drynaria acuminata (Willdenow) C. V. Morton is an epilithic fern belonging to the subfamily Drynarioideae of Polypodiaceae (PPG I Citation2016). This species is rather widespread in Indonesia, Malaysia, and the Philippines with only a few occurrences in Cambodia, Laos, Thailand, and Vietnam recorded whilst it was only recorded to occur in China till 1990 (Li Citation1990). The only Chinese location is in Xishuangbanna, Yunnan and is considered to be endangered by deforestation and loss of habitat. Thus, the species was included into the Red List of China Higher Plants (Qin et al. Citation2017). Based on the herbarium record, D. acuminata is considered here as a plant species with extremely small populations (PSESP) in China (Ma et al. Citation2013). In the past, this species was treated as the monotypic genus Photinopteris J.Sm. (Zhang and Gilbert Citation2013) but it was later transferred to Aglaomorpha (PPG I Citation2016) and now to Drynaria nom. consv. (Christenhusz and Schneider Citation2012).
In this study, we reported the complete chloroplast genome of D. acuminata for the first time. Fresh leaf material was collected from Xishuangbanna Tropical Botanical Garden, Chinese Academy of Science, Yunnan, China (N21°55′44″, E101°15′20″) and the voucher specimen was deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden (HITBC, collection number: Liu-CP13). Genomic DNA was extracted from 2 g leaves using the CTAB method (Doyle and Dickson Citation1987), 0.5 µg DNA was fragmented to reconstruct short-insert (500 bp) libraries following the manufacturer’s manual (Illumina) and then used for sequencing. The DNA sample was indexed by tags and pooled together in one lane of a Genome Analyzer (Illumina HiSeq 2000) for sequencing at the Germplasm Bank of Wild Species, Kunming Institute of Botany (KIB) in Kunming, China. GetOrganelle toolkit (Jin et al. Citation2020) and Geneious (https://www.geneious.com) were employed to assemble and annotate the genome. CP genome of Pyrrosia petiolosa (MN885667) and Drynaria roosii (KY075853) was employed as reference genome The newly sequenced and annotated plastid genome was submitted to the GenBank (accession number MW042681).
The chloroplast genome of D. acuminata had the typical quadripartite structure; the total length is 151,591 bp including a large single-copy (LSC) region of 80,661 bp, a small single-copy (SSC) region of 21,688 bp, and a pair of inverted repeats (IR) regions of 24,621 bp. The chloroplast genome contains 131 genes including 88 protein-coding genes, 35 tRNAs, and 8 rRNA genes.
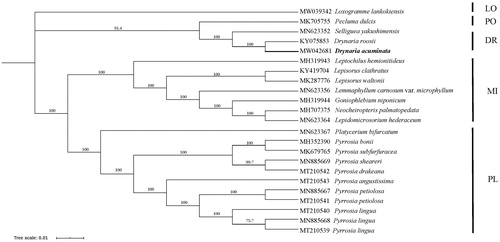
The chloroplast genome of D. acuminata was incorporated into a matrix including 23 taxa covering four subfamilies of Polypodiaceae (PPG I Citation2016) with the aim to reconstruct phylogenetic relationship of Polypodiaceae. 84 coding genes were selected, aligned, and concatenated into a single matrix using MAFFT (Katoh and Standley Citation2013). The phylogenetic analyses were constructed using IQ-tree with Maximum likelihood method (Minh et al. Citation2020), nucleotide substitution model of K-2-P was used with the 1000 bootstrap replicates. Loxogramme lankokiensis was selected as outgroup according to PPG I (Citation2016).
In the phylogenetic tree, D. acuminata was grouped together with D. roosii with Selliguea yakushimensis and Pecluma dulcis formed a basal clade (). Two subfamilies (Microsoroideae and Platycerioideae) were strongly supported as monophyletic and were found to be sister clades. Subfamily Polypodioideae was founded to be clustered with Drynarioideae. All Pyrrosia species were grouped together and formed a monophyletic clade.
Figure 1. Maximum likelihood phylogeny reconstructed from 23 chloroplast genomes by IQ-tree. The sampling covered representatives of four subfamilies of Polypodiaceae. Loxogramme lankokiensis was selected as outgroup. DR: Drynarioideae; PO: Polypodioideae; MI: Microsoroideae; PL: Platycerioideae; LO: Loxogrammoideae.
The complete chloroplast genome sequence of D. acuminata will provide useful information for phylogeography of this tropical fern species and the phylogenomic study for the derived fern family Polypodiaceae.
Disclosure statement
The author is responsible for the content and no potential conflict of interest among the authors.
Data availability statement
The data that support the findings of this research are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW042681.
Additional information
Funding
References
- Christenhusz MJM, Schneider H. 2012. Proposal to conserve the name Drynaria against Aglaomorpha (Polypodiaceae). Taxon. 61(2):465–466.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNArestriction endonuclease analysis. Taxon. 36(4):715–722.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li QJ. 1990. Photinopteris J. Sm., a new recorded genus of Drynariaceae in China. J Syst E. 28(5):412–413.
- Ma YP, Chen G, Grumbine RE, Dao ZL, Sun WB, Guo HJ. 2013. Conserving plant species with extremely small populations (PSESP) in China. Biodivers Conserv. 22(3):803–809.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von HA, Lanfear R. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Syst E. 54:563–603.
- Qin HN, Yang Y, Dong SY, He Q, Jia Y, Zhao LN, Yu SX, Liu B, Yan YH, Xiang JY, et al. 2017. Threatened species list of China’s higher plants. Biodiv Sci. 25(7):696–744.
- Zhang XC, Glibert MG. 2013. Photinopteris. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 766.
