Abstract
The complete chloroplast genome (plastome) of the Chenopodium acuminatum was assembled and annotated in this study. The complete plastome was composed of circular DNA molecules with a total length of 152,200 bp, comprising a large single-copy region (83,683 bp), a small single-copy region (18,131 bp), and two inverted repeat regions (25,193 bp). GC content of this complete plastome was 37.2%. In total, 113 unique genes were annotated, including 79 protein-coding genes (PCGs), 30 transfer RNAs, and 4 ribosomal RNAs. Phylogenomic analysis showed that C. acuminatum was closely related to C. album.
The genus Chenopodium sensu lato, comprising ca. 150 species, belongs to the subfamily Chenopodioideae (Amaranthaceae, Caryophyllales), mainly distributed in temperate and subtropical regions of the world (Hong et al. Citation2017). Chenopodium is one of the largest genera in Chenopodioideae subfamily with complex taxonomy due to highly polymorphic leaf shape, floral structure, and seed morphology shared by many Chenopodium species (Rahiminejad and Gornall Citation2004; Kurashige and Agrawal Citation2005). These are mostly herbaceous, suffrutescent, and arborescent perennial species which were found in arid/semi-arid regions, and saline-alkali soils (Yao et al. Citation2019). Species of this genus lack typical adaptive structures such as the Kranz type leaf anatomy, the C4 photosynthetic pathway, and succulence when compared to other plants in dry environments (Fuentes-Bazan et al. Citation2012; Qu, Li, et al. Citation2019; Zhang et al. Citation2019). Chenopodium species have fascinated many scientists because of their high nutritional value as well source of leafy vegetables worldwide. Furthermore, it contains many bioactive compounds, such as flavonoids, volatile components, saponins and phenolic acids, etc., which is used in herbalism and pharmaceutical industries (Tang and Tsao Citation2017). Chenopodium acuminatum Willd. is an annual herb, mainly distributed in China, Japan, and Korea. C. acuminatum is a hermaphrodite plant, pollinated by wind, and generally found along riverbanks, wastelands, and field margins. Plastome has been used to resolve the phylogenetic relationship and development of molecular makers for the identification of taxonomically complex plant species (Jansen et al. Citation2007; Huang et al. Citation2020). In this study, we assembled the complete plastome of C. acuminatum, in order to facilitate genomic resources to provide insight into systematic and evolution of this important species.
Chenopodium acuminatum fresh leaves were collected from Changqing District (Shandong, China; 36°32′N, 116°50′E) and voucher specimen (SD124) was deposited at College of Life Sciences, Shandong Normal University, China. Total genomic DNA was extracted by the modified CTAB method, and was used for library preparation and genome sequencing using Illumina MiSeq at Novogene (Beijing, China). Plastome assembly was conducted by Organelle Genome Assembler (OGA) as described in Qu, Fan, et al. (Citation2019). Plastid Genome Annotator (PGA, (Qu, Moore, et al. Citation2019)) was used to annotate the complete plastome. The raw sequencing reads was deposited in SRA with the accession number SRR12894192, and the chloroplast genome was deposited in the GenBank with the accession number MW057780.
The complete plastome of C. acuminatum was quadripartite structure with 152,200 bp in length, comprising a large single-copy region (83,683bp), a small single-copy region (18,131bp), and a pair of inverted repeat regions (25,193bp). GC content of this complete plastome was 37.2%. A total of 113 unique genes were annotated, including 79 protein-coding genes (PCGs), 30 transfer RNAs, and 4 ribosomal RNAs, among which 13 PCGs and 8 transfer RNAs contained introns.
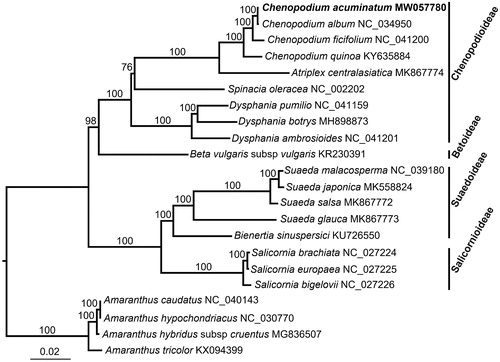
Phylogenetic relationship of C. acuminatum within the Amaranthaceae family was inferred using previously published seventeen plastomes and Amaranthus caudatus, A. hypochondriacus, A. hybridus subsp. cruentus and A. tricolor were used as outgroups. Maximum-likelihood tree was reconstructed by RAxML v8.2.10 (Stamatakis Citation2014), based on 79 shared protein-coding genes aligned by MAFFT v7.313 (Katoh and Standley Citation2013), and with 1000 rapid bootstrap replicates and GTRGAMMA substitution model. Phylogenomic analysis showed that C. acuminatum was closely related to C. album (). This C. acuminatum plastome will provide a genomic resources for future systematic studies of this complex genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov/, SRA accession number SRR12894192 and GenBank accession number MW057780.
References
- Fuentes-Bazan S, Mansion G, Borsch T. 2012. Towards a species level tree of the globally diverse genus Chenopodium (Chenopodiaceae). Mol Phylogenet Evol. 62(1):359–374.
- Hong SY, Cheon KS, Yoo KO, Lee HO, Cho KS, Suh JT, Kim SJ, Nam JH, Sohn HB, Kim YH. 2017. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album. Front Plant Sci. 8: 1696.
- Huang R, Liang Q, Wang Y, Yang TJ, Zhang Y. 2020. The complete chloroplast genome of Epimedium pubescens Maxim. (Berberidaceae), a traditional Chinese medicine herb. Mitochondrial DNA B. 5(3):2042–2044.
- Jansen RK, Cai ZQ, Raubeson LA, Daniell H, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kurashige NS, Agrawal AA. 2005. Phenotypic plasticity to light competition and herbivory in Chenopodium album (Chenopodiaceae). Am J Bot. 92(1):21–26.
- Qu XJ, Fan SJ, Wicke S, Yi TS. 2019. Plastome reduction in the only parasitic gymnosperm Parasitaxus is due to losses of photosynthesis but not housekeeping genes and apparently involves the secondary gain of a large inverted repeat. Genome Biol Evol. 11(10):2789–2796.
- Qu XJ, Li XT, Zhang LY, Zhang XJ, Fan SJ. 2019. Characterization of the complete chloroplast genome of Suaeda salsa (Amaranthaceae/Chenopodiaceae), an annual succulent halophyte. Mitochondrial DNA B. 4(2):2133–2134.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50
- Rahiminejad MR, Gornall RJ. 2004. Flavonoid evidence for allopolyploidy in the Chenopodium album aggregate (Amaranthaceae). Plant Syst E. 246(1-2):77–87.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tang Y, Tsao R. 2017. Phytochemicals in quinoa and amaranth grains and their antioxidant, anti-inflammatory, and potential health beneficial effects: a review. Mol Nutr Food Res. 61(7):1600767.
- Yao Y, Li XT, Wu XY, Fan SJ, Zhang XJ, Qu XJ. 2019. Characterization of the complete chloroplast genome of an annual halophyte, Chenopodium glaucum (Amaranthaceae). Mitochondrial DNA B. 4(2):3898–3899.
- Zhang XJ, Wang N, Zhang LY, Fan SJ, Qu XJ. 2019. Characterization of the complete plastome of Atriplex centralasiatica (Chenopodiaceae), an annual halophytic herb. Mitochondrial DNA B. 4(2):2475–2476.