Abstract
Sansevieria trifasciata var. laurentii (S. trifasciata) is a kind of popular in-door and out-door plant around world, it is not only known as the ornamental plant, but also as medical plant. It belongs to the Draceanaceae family, Draceanaceae includes more than 60 species distributed in tropical and subtropical dry climate regions. In this study, we sequenced the sample of S. trifasciata and determined its complete chloroplast genome. The length of CP genome is 155,179 bp, includes two invert repeats (IR) regions of 26,513 bp, a large single copy (LSC) region of 83,680 bp and a short single copy (SSC) region of 18,473 bp. There are 133 genes, which includes 87 protein coding genes, 8 rRNA and 38 tRNA, and 37.5% overall GC content. Each of trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA genes contains a intron, clpP and ycf3 contains 2 intron. The phylogenetic position shows that S. trifasciata has the closest relationship with Rohdea Chinensis (MH356725.1).
Sansevieria trifasciata var. laurentii (S. trifasciata) is a kind of popular in-door and out-door plant around world, however S. trifasciata is not only known as the ornamental plant, but also as medical plant. In traditional medical care, it was used for antitussive and an expectorant, and in South America it was sold as a crude drug in the market to treat victims of cough, snake bite, sprain, bruise, boil, abscess, respiratory inflammation and hair tonic(Stover Citation1983; Antunes et al. Citation2003) Furthermore, S. trifasciata is a kind of raw sauce of medicine (saponins) that can be used for tumor cell growth inhibitor and lowering blood cholesterol level. That medicine can also be used as spermicide (contraception), anti-inflammation, cytotoxic and antimicrobial agents (Dey Citation2014; Odeh and Amom Citation2014). Sansevieria trifasciata belongs to the Draceanaceae family, which includes more than 60 species distributed in tropical and subtropical dry climate regions (Lu and Morden Citation2014). Base the situation that no complete chloroplast genome of the species in Sansevieria has been published, so in this study we sequenced the sample of S. trifasciata and determined its complete chloroplast genome.
The sample of S. trifasciata was collected from South China Botanical Garden, Tianhe District, Guangzhou, Guangdong Province (N113°22′34ʺ, E23°11′32ʺ). We used the fresh leaves to extract chloroplast DNA based CTAB method (Doyle and Doyle Citation1987) and constructed the libraries with an average length of 350 bp using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA). Then the libraries were sequenced on Illumina Novaseq 6000 platform, over 2 Gb clean data was assembled by SPAdes v.3.11.0 software (Bankevich et al. Citation2012) and annotated by GeSeq (Tillich et al. Citation2017). The complete sequence and annotation results were submitted to GenBank, under the accession number (MT922036) and the sample was stored at Laboratory of Molecular Biology, Liaocheng University, Liaocheng (Voucher specimen: ST20200701LP).
The complete chloroplast genome of S. trifasciata is 155,179 bp in length, and contains a large single copy (LSC) with 83,680 bp in length, a small single copy (SSC) with 18,473 bp in length and two inverted repeat (IR) regions of 26,513 bp each. There were 133 genes, which includes 87 protein coding genes, 8 rRNA and 38 tRNA, and 37.5% overall GC content. Each of trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA genes contains a intron, clpP and ycf3 contains 2 introns.
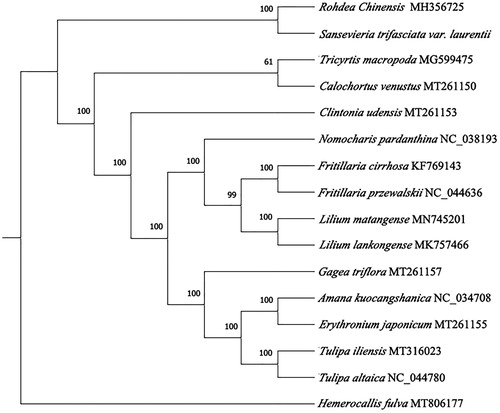
To confirm the phylogenetic position and understand the relationship of S. trifasciata. The complete chloroplast genome of 15 species were collected and aligned with S. trifasciata by MAFFT7.037 (Katoh and Standley Citation2013). Subsequently, the phylogenetic tree was constructed by IQTREE v1.6 (Nguyen et al. Citation2015; Hoang et al. Citation2018) with 1000 bootstraps replicates using Best-fit model. By using Hemerocallis fulva (LC554221.1) as out group we got the final ML tree, then showed that S. trifasciata had the closest relationship with Rohdea chinensis (MH356725.1).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The sequencing data that support the finding of this study are openly available in NCBI Sequence Read Archive(SRA) with accession number: SRR12980945. The assembled complete chloroplast genome sequence of Sansevieria trifasciata var. laurentii has been submitted to GenBank under the accession number: MT922036 (https://www.ncbi.nlm.nih.gov/nuccore/MT922036.1).
Additional information
Funding
References
- Antunes AD, Da Silva BP, Parente JP, Valente AP. 2003. A new bioactive steroidal saponin from Sansevieria cylindrica. Phytother Res. 17(2):179–182.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dey B, Bhattacharjee R, Rajeev A, Mitra A, Singla K. 2014. Mechanistic explorations of antidiabetic potentials of Sansevieria trifasciata. Indo Global J Pharmaceut Sci. 4(2):113–122.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lu P-L, Morden CW. 2014. Phylogenetic relationships among dracaenoid genera (Asparagaceae: Nolinoideae) inferred from chloroplast DNA loci. Syst Bot. 39(1):90–104.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Odeh IC, Amom TAT. 2014. Phytochemical and antimicrobial evaluation of leaf-extracts of Pterocarpus santalinoides. Euro J Med Plants. 4(1):105–115.
- Stover H. 1983. Sansevieria book. 1st ed. California: Endangered Species Press.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.