Abstract
In the present study, we obtained the first complete mitochondrial genome sequence of Leucosoma (Salanx) chinensis. It was 16,595 bp in length and consisted of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a non-coding control region. The nucleotide composition was A (23.1%), T (25.2%), G (20.0%) and C (31.8%), and the A + T content (48.3%) was a little lower than G + C content (51.80%). Phylogenetic analysis of 10 species of Salangidae identified three major clades. These results may facilitate the future genetic research of L. chinensis and Salangidae.
The Chinese noodlefish Leucosoma chinensis (Salangidae, Osmeriformes, Osteichthyes) is endemic to east and south Chinese seas (Zheng Citation1989). It is sometimes classified in the genus Salanx (e.g. Froese and Pauly Citation2019). It has a slender body, a very flat head, a long snout, and a small fleshy protrusion on the front of the lower jaw. Each fin is small, the dorsal fin is located above the anal fin, and there is a pair of small transparent adipose fins between the dorsal and caudal fins. Its body is soft and transparent without scales. It usually lives in coastal waters, and reproduces in brackish or fresh water during the reproductive period. It mainly feeds on zooplankton. Members of this species are small, and the meat is considered delicious and nutritious (Su and Wang Citation1985; Zheng Citation1989; Huang et al. Citation2012). In the present study, we obtained the first complete mitochondrial genome sequence of L. chinensis, which may facilitate future genetic research on this species.
A sample of L. chinensis was collected from the Wuzhou (N23°28′23″, E111°22′16″) section of the Xijiang River in 2019. The fish was placed in 95% alcohol and reposited in a refrigerator at −20 °C in the specimen room of the School of Life Sciences, Jianghan University (Sample code: L. chinensis 20190712001). Total genomic DNA from the dorsal muscle was extracted using the Foregene Animal Tissue DNA Kit. The complete mitochondrial genome was obtained from Illumina high-throughput sequencing and de novo assembled using NOVOPlasty ver 2.6 (Dierckxsens et al. Citation2017). The tRNA genes were predicted using the program ARWEN (Laslett and Canback Citation2008), rRNA and protein-coding genes were created using web server DOGMA (Wyman et al. Citation2004).
The complete mitochondrial genome of L. chinensis was 16,595 bp in length (GenBank accession number: MW131880). The nucleotide composition was A (23.1%), T (25.2%), G (20.0%) and C (31.8%), and the A + T content (48.3%) was a little lower than G + C content (51.80%). The complete mitochondrial genome consisted of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a non-coding control region. ND6 gene and 8 tRNA genes (tRNAPro, tRNAGlu, tRNASer, tRNATyr, tRNACys, tRNAAsn, tRNAAla and tRNAGln) were encoded on the light strand, others were encoded on the heavy strand.
Following Sangster and Luksenburg (Citation2020), we verified the identity and integrity of our mitogenome sequence of L. chinensis with reference sequences of three commonly used markers in fish systematics: NADH dehydrogenase subunit 1 (ND1, 975 bp; 63 sequences of Salangidae, incl. three of L. chinensis), part of cytochrome oxidase subunit I (COI, 655 bp; 193 sequences of Salangidae, incl. five of L. chinensis), and cytochrome b (Cyt b, 1141 bp; 307 sequences of Salangidae, incl. three of L. chinensis).
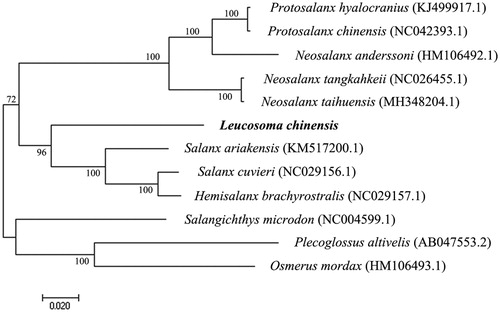
In order to explore the evolutionary status of L. chinensis within the Salangidae, a Neighbor-Joining phylogenetic tree was constructed by MEGA7 (Kumar et al. Citation2016) based on the complete mitochondrial genome sequences of 10 species of Salangidae and two species (Plecoglossus altivelis, Osmerus mordax) as the outgroup (). Except for L. chinensis, mitochondrial genome sequences of other species were downloaded from the NCBI GenBank. The phylogenetic tree showed that the 10 species of Salangidae clustered into three major clades: Protosalanx hyalocranius, Protosalanx chinensis, Neosalanx anderssoni, Neosalanx tangkahkeii, and Neosalanx taihuensis in clade1, Leucosoma chinensis, Salanx ariakensis, Salanx cuvieri, and Hemisalanx brachyrostralis in clade2, and Salangichthys microdon in clade3. The phylogenetic relationships inferred in our study based on mitogenome sequences were very similar to those obtained in a previous study based on cytochrome b sequences (Zhang et al. Citation2007).
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW131880.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Froese R, Pauly D, editors. 2019. FishBase. World Wide Web electronic publication. www.fishbase.org. version (12/2019).
- Huang XY, Zhang Q, Si CL, Ma B. 2012. Genetic diversity of Leucosoma chinensis based on the cyt b gene. Jiangsu Agric Sci. 40(4):45–48. (in Chinese).
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Sangster G, Luksenburg JA. 2020. The published complete mitochondrial genome of Eptesicus serotinus is a chimera of Vespertilio sinensis and Hypsugo alaschanicus (Mammalia: Chiroptera). Mitochondrial DNA Part B. 5(3):2661–2664.
- Su YQ, Wang J. 1985. Food and eating habits of Salanx acuticeps in the Jiulong River Estuary. J Oceanography Taiwan Strait. 4(2):219–226. (in Chinese).
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20(17):3252–3255.
- Zhang J, Li M, Xu M, Takita T, Wei F. 2007. Molecular phylogeny of icefish Salangidae based on complete mtDNA cytochrome b sequences, with comments on estuarine fish evolution. Biol J Linn Soc 91(2):325–340.
- Zheng C, editor. 1989. The fishes of Pearl River. Beijing: Science Press (in Chinese).