Abstract
Dayu yak (Bos grunniens) is a long-furred yak breed from the Qinghai-Tibetan Plateau, and is highly adapted to local high-altitude and cold environments. In this study, its mitochondrial genome was characterized via high-throughput sequencing technology. The genome is 16,323 bp long with an AT-biased base composition (61.0% A + T; light strand), and harbors the typical set of 37 mitochondrial genes and a noncoding control region. Its gene arrangement is identical to those of other bovid taxa. Phylogenetic analysis suggests that Dayu yak is most closely related to Maiwa, Niangya, Qinghai Plateau, Xueduo and Yushu yaks.
As an iconic symbol of the Qinghai-Tibetan Plateau, domestic yaks (Bos grunniens) have long been exploited for meat, milk, transportation and other necessities by local communities (Qiu et al. Citation2012). Dayu yak is a long-furred yak breed with its distribution mostly restricted to Haibei Tibetan Autonomous Prefecture, Qinghai Province, China, and is highly adapted to local high-altitude and cold environments. To facilitate its genetic assays, its complete mitochondrial genome was assembled and annotated via high-throughput sequencing technology in this study. Furthermore, phylogenetic analysis was also conducted to ascertain its relationship with other taxa within the subfamily Bovinae. The annotated genomic sequence has been deposited into GenBank under the accession number MT649465.
The blood sample of Dayu yak were collected from Yeniugou Township, Qilian County, Haibei Tibetan Autonomous Prefecture, Qinghai Province, China (38.35°N, 99.34°E). The voucher specimen (DAYU20200528) is stored in the Key Laboratory of Yak Breeding Engineering of Gansu Province, Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS (Lanzhou, Gansu Province, China). Total genomic DNAs were isolated and purified using the QIAamp DNA Blood Mini Kit (Qiagen, CA, USA). Following the library preparation, high-throughput sequencing was carried out with the Illumina HiSeq XTM Ten Sequencing System (Illumina, CA, USA) by Annoroad Gene Technology (Beijing, China). Totally, 16.03 M raw reads of 150 bp were obtained, and were used to assemble the mitochondrial genome with MITObim v1.9 (Hahn et al. Citation2013); the reference sequence (JQ692071) was previously published by Qiu et al. (Citation2012). Annotation of the mitochondrial genome was done in Geneious R11 (Biomatters Ltd., Auckland, New Zealand) by comparing with those of other bovid taxa.
The mitochondrial genome of Dayu yak is 16,323 bp long with an A + T-biased base composition (33.7% A, 25.8% C, 13.2% G and 27.3% T; ‘light strand’), and harbors the typical set of 37 animal mitochondrial genes (13 protein-coding genes, 22 tRNAs and two rRNAs) and one non-coding control region. Its gene arrangement is identical to those of other bovid taxa. The PCGs start with the typical ATA (ND2, ND3 and ND5) or ATG (the 10 other PCGs) initiation codons, and end with TAG (ND2), the incomplete T (COX3, ND3 and ND4) or TAA (the nine others) termination codons. The tRNAs range in size from 60 (tRNA-SerAGN) to 75 bp (tRNA-LeuUUR) with a total length of 1509 bp. The two rRNAs are 957 bp (12S rRNA) and 1,571 bp (16S rRNA) long, respectively, and are separated by tRNA-Val. The control region is 893 bp long, and is present between tRNA-Pro and tRNA-Phe. Besides, a 31-bp-long origin of the L-strand replication is identified between tRNA-Asn and tRNA-Cys.
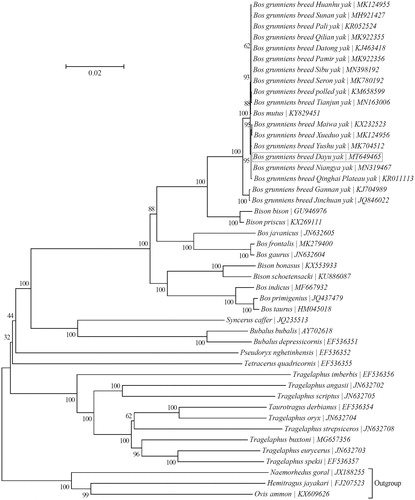
To investigate the relationship of Dayu yak with 42 other taxa within the subfamily Bovinae, a neighbor-joining phylogenetic tree was reconstructed using the concatenated sequences of all 13 protein-coding genes (alignment size: 11,370 bp) with MEGA7 (http://www.megasoftware.net/) (Hall Citation2013; Kumar et al. Citation2016) (). Bootstrap support values were estimated from 1000 random samplings. The outgroup taxa included in the phylogenetic analysis are three species from the subfamily Caprinae, i.e. Hemitragus jayakari (FJ207523) (Hassanin et al. Citation2009), Naemorhedus goral (JX188255) (Yang et al. Citation2013) and Ovis ammon (KX609626) (Mao et al. Citation2017). The result suggests that Dayu yak is more closely related to five local yak breeds (Maiwa, Niangya, Qinghai Plateau, Xueduo and Yushu yaks) than to the other 37 taxa within the subfamily Bovinae ().
Figure 1. Phylogeny of the subfamily Bovinae based on the neighbor-joining analysis of the concatenated sequences of 13 mitochondrial protein-coding genes (alignment size: 11,370 bp). The bootstrap values next to the nodes are based on 1000 random samplings. Three species within the subfamily Caprinae were included as outgroup taxa.
Disclosure statement
The authors declare that there are no financial and personal relationships with other people or organizations that can an appropriately influence our work, and no professional or other personal interest of any nature or kind in any product, service and company that could be construed as influencing the position presented in, or the review of the manuscript.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT649465.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 30(5):1229–1235.
- Hassanin A, Ropiquet A, Couloux A, Cruaud C. 2009. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol. 68(4):293–310.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Mao H, Liu H, Ma G, Yang Q, Guo X, Lamaocao Z. 2017. The complete mitochondrial genome of Ovis ammon darwini (Artiodactyla: Bovidae). Conserv Genet Resour. 9(1):59–62.
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44(8):946–949.
- Yang C, Xiang C, Qi W, Xia S, Tu F, Zhang X, Moermond T, Yue B. 2013. Phylogenetic analyses and improved resolution of the family Bovidae based on complete mitochondrial genomes. Biochem Syst Ecol. 48:136–143.
