Abstract
Medicago archiducis-nicolai Sirj. is a well-known high-quality forage as its good palatability and strong tolerance to drought, cold and saline-alkali stress. Here, the complete chloroplast genome sequence of M. archiducis-nicolai was reported. The size of the complete chloroplast genome is 127,072 bp in length. The chloroplast genome has no inverted repeat (IR) regions, which is very common in the family Fabaceae. The M. archiducis-nicolai chloroplast genome encodes 106 genes: 72 protein-coding genes, 30 tRNAs, and 4 rRNAs. The phylogenetic analysis result strongly suggested that M. archiducis-nicolai is a distinct lineage in Medicago, being sister to highly supported clade composed of three species (M. hybrida, M. papillosa and M. sativa).
Medicago archiducis-nicolai Sirj. is a perennial plant of Medicago, which is one of the core species of Section Pialcarpae (Small and Marcel Citation1989). Medicago archiducis-nicolai is mainly distributed in Qinghai, Tibet, and adjacent to high-altitude areas. It is the only perennial wild alfalfa that can survive the winter in the natural alpine grassland of Qinghai Tibet Plateau (Jin et al. Citation2013). It is widely distributed in the areas with an altitude of 1500–4250m (Li et al. Citation1997; De and Xu Citation2009), and grows better in the sunny slope grassland, river beach and secondary bare land. It has strong tolerance to drought, cold and saline-alkali stress (Balabaev Citation1934). Compared with Medicago ruthenica, M. archiducis-nicolai as an indigenous species in the Tibetan Plateau, has a more severe growth environment and stronger adaptability to extreme environments (Wang et al. Citation2020). At the same time, M. archiducis-nicolai is a perennial wild legume species, has high crude protein content and good palatability (Dekejia and Xu 2009) has dense underground rhizomes with rich nodules, and also has strong barren and trampling resistance (Liu Citation1987; Li Citation2007a). For these reasons, M. archiducis-nicolai is considered to be a high-quality perennial leguminous forage resource with domestication potential, which is expected to be cultivated and utilized in areas where M. sativa and other alfalfa species cannot survive the winter (Li Citation2007b). However, no studies on the plastome of M. archiducis-nicolai have been published. In this study, the complete chloroplast genome of M. archiducis-nicolai (Genbank accession number: MN901634) was sequenced on the Illumina NovaSeq Platform (Illumina, San Diego, CA, USA), which will provide genetic and genomic information to promote its ecological restoration and systematics research of Fabaceae. The raw data was submitted on Sequence Read Archive (Number: SRR12951164).
In this study, M. archiducis-nicolai were collected from Garang village, Guide County, Haidong City, Qinghai Province, China (36.32°N, 101.52°E). The fresh and young leaves were dried immediately by silica gels. The complete chloroplast genome of M. archiducis-nicolai was extracted from the dried leaves (about 0.2 g) with a modified CTAB method. The voucher specimen was kept in the Herbarium of the Northwest Institute of Plateau Biology, Chinese Academy of Sciences (HNWP, XIE2019014). Genome sequencing was performed using the Illumina NovaSeq Platform at Genepioneer Biotechnologies Inc., Nanjing, China. The trimmed reads were mainly assembled by SPAdes (Bankevich et al. Citation2012). Then, PCGs, rRNAs and tRNAs were annotated by blast v2.2.25, hmmer v3.1b2 and aragorn v1.2.38, respectively.
The complete chloroplast genome of M. archiducis-nicolai has an atypical chloroplast genome structure with a length of 127,072 bp. This chloroplast genome has no inverted repeat (IR) regions, which is very common in the family Fabaceae. The GC content of the whole chloroplast genome is 34.23%. A total of 106 functional genes were annotated, including 72 protein-coding genes (PCGs), 30 tRNA genes and 4 rRNA genes. 14 of them contain 1 intron and 1 of them contains 2 introns.
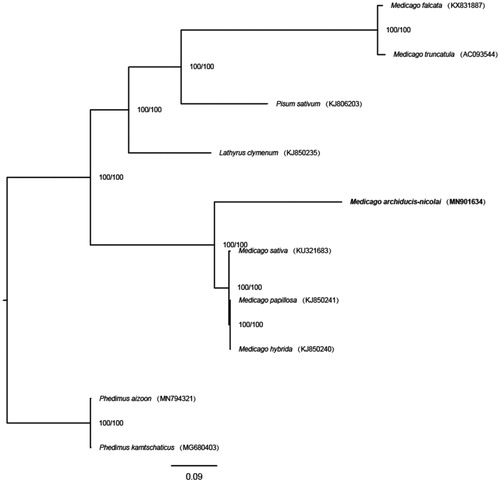
Eight complete chloroplast genomes of Fabaceae (the number from Medicago and Pisum are 7 and 1, respectively) and two outgroups (two species from Crassulaceae Phedimus) were used for constructing maximum likelihood with 1000 bootstrap repeats (model: K3Pu + F) by W-IQ-TREE (Trifinopoulos et al. Citation2016) after aligned by MAFFT 7 (Katoh and Standley Citation2013) (). The phylogenetic tree showed that M. archiducis-nicolai was sister to the clade composed of three species (M. hybrida, M. papillosa and M. sativa).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov (Reference number: MN901634) and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/search/all/?term=SRR12951164 (Number: SRR12951164).
Additional information
Funding
References
- Balabaev GA. 1934. Yellow lucernes of Siberia, Medicago ruthenica (L.) Lebd. and M. platycarpos (L.) Lebd. Bull App Bot Genet Plant Breed Serv. 7:13–123.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikow AS, Lesin VN, Nikloenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- De KJ, Xu CT. 2009. Introduction and domestication of Medicago archiducis-nicolai, a natural legume forage in alpine region. Seeds. 28(7):73–75.
- Jin D, Ma J, Ma W, Liang C, Shi Y, He JS. 2013. Legumes in Chinese natural grasslands: species, biomass, and distribution. Rangeland Ecol Manage. 66(6):648–656.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li J, Zhang QB, Yang G. 1997. Medicago in china and the resource superiority in Xinjiang. Acta Agrestia Sinica. 5(4):286–291.
- Li YL. 2007a. Effect of rhizobium inocultion on Medicago archiducis-nicolai Sirj. Chin Qinghai J Vet Sciences. 37(4):5–6.
- Li YL. 2007b. Observation and study of aboveground biomass of Medicago archiducis-nicolai Sirj in Xining Area. Prataculture Anim Husbandry. 142:16–17.
- Liu YT. 1987. A fine wild legume grass in Qinghai and Tibet, Medicago archiducis-nicolai. Chin J Grassland. 03:71–73.
- Small E, Marcel J. 1989. A synopsis of the genus Medicago (Leguminosae). Can J Bot. 67(11):3260–3294.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–235.
- Wang YF, Zhang YM, Liu DM, Shen YF, Wang HQ. 2020. Development and verification of EST-SSR markers in Medicago archiducis-nicolai by transcriptome sequencing. Pratacult Sci. 37(4):718–727.