Abstract
Carya hunanensis W. C. Cheng & R. H. Chang is a tree of great economic importance in China. In this study, the complete chloroplast genome of C. hunanensis was determined and analyzed phylogenetically. The whole genome was determined to be 159,892 bp in length, presenting a typical quadripartite structure with a large single copy (LSC) region (89,466 bp), a small single copy (SSC) region (18,714 bp), and a pair of inverted repeat (IR) regions (25,856 bp each). There were 132 genes annotated in the genome, including 84 protein-coding genes, 40 tRNA genes, and eight rRNA genes. The overall GC content of C. hunanensis chloroplast genome was 36.24%. Phylogenetic analysis indicated that C. hunanensis was closely related to C. kweichowensis and Annamocarya sinensis.
Carya hunanensis is an economic important tree species with straight trunk and good wood quality. It possesses excellent drought resistance and good adaptability to poor soil environments (Fan et al. Citation2007). The kernel of this species contains a relative high content of linoleic acid, linolenic acid, and arachidonic acid, which are the essential fatty acids in humans (Fan et al. Citation2007; Peng et al. Citation2012). Besides, its kernel includes a total of 16 amino acids, of which nine are essential for the nutrition of man (Fan et al. Citation2007). Nowadays, there is still genomic data regarding C. hunanensis. Chloroplast genome is a circular double-stranded DNA molecule, with size ranging from 120 kb to 150 kb, and generally have a low mutation rate, making it suitable for phylogenetic and population genetic studies (Daniell et al. Citation2016; Xu et al. Citation2020; Yang et al. Citation2020). In the present study, we assembled the complete chloroplast genome of C. hunanensis, and analyzed its phylogenetic position.
Fresh and healthy leaves were collected from the experimental farm of Nanjing Botanical Garden (Nanjing, China, 118°51′12.73″E, 32°2′11.28″N) and were deposited at Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (voucher specimen no. Zhai20200711-1). Genomic DNA was isolated using a Universal Plant Total DNA Extraction Kit (BioTeke, Beijing, China) according to the manufacturer's protocol. A paired-end library with approximate insert lengths of 300 bp was built using an Illumina Hiseq Library Kit (Illumina, San Diego, CA). The resulting paired-end library was sequenced on an Illumina Hiseq 4000 platform (Illumina, San Diego, CA). After fulfilling sequencing, approximately 34.42 M raw reads were generated (NCBI SRA accession number SRR12771063), and were further filtered by Trimmomatic v0.32 (Bolger et al. Citation2014) to obtain clean reads. Assembly the chloroplast genome was conducted by NOVOPlasty (Dierckxsens et al. Citation2017), and annotation was performed by Geneious R11 v11.0.5 (Biomatters Ltd., Auckland, New Zealand) with C. illinoinensis as a reference (MN977124; Mo et al. Citation2020). The annotated cp genome was deposited in GenBank (accession number MT955359).
The entire chloroplast genome of C. hunanensis was 159,892 bp in size, exhibiting a conserved quadripartite structure. It contained pair of inverted repeats (IRa and IRb) of 25,856 bp, a large single copy (LSC) region of 89,466 bp, and a small single copy (SSC) region of 18,714 bp. The chloroplast genome of C. hunanensis was predicted to possess a total of 132 genes, including 84 protein-coding genes, 40 tRNA genes, and eight rRNA genes. For the 132 genes, there were six protein-coding genes, seven tRNA genes, and four rRNA genes duplicated in the IR regions. The overall GC content in the chloroplast genome of C. hunanensis was 36.24%, and the corresponding values for the LSC, SSC, and IR regions were 33.83%, 30.03%, and 42.66%, respectively.
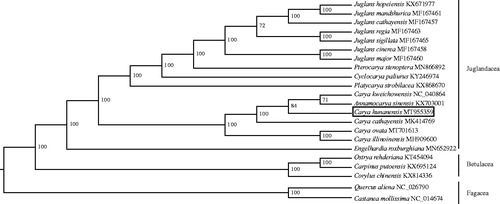
To examine the phylogenetic status of C. hunanensis, phylogenetic analysis was conducted using the complete chloroplast genome sequences of 22 species: 17 from Juglandacea, three from Betulacea, and two from Fagacea. Multiple sequence alignment was performed by MAFFT program (Katoh and Standley Citation2013), and the aligned sequences were used to construct phylogenetic tree by IQ-tree program (Nguyen et al. Citation2015) with the maximum-likelihood method. Previously, Annamocarya sinensis was reported to be a synonym of Carya sinensis. Besides, the molecular analysis indicated that A. sinensis was a representative of the Asian Carya species (Zhang et al. Citation2013; Mo et al. Citation2020). Our phylogenetic analysis () showed that the Asian Carya species: C. kweichowensis, A. sinensis, C. hunanensis, and C. cathayensis form a paraphyletic clade, and C. hunanensis had a close relationship with A. sinensis and C. kweichowensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MT955359. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA667312, SRR12771063, and SAMN16365445, respectively.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fan W, An H, Long L, Ma W, Yang W. 2007. Study on nutrient component in kernel of Carya hunanensis Cheng. Chin Wild Plant Resour. 26(5):64–65.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Mo Z, Lou W, Chen Y, Jia X, Zhai M, Guo Z, Xuan J. 2020. The chloroplast genome of Carya illinoinensis: genome structure, adaptive evolution, and phylogenetic analysis. Forests. 11(2):207.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Peng X, Fu H, Yang C. 2012. Study on extracting technology and properties of Carya hunanensis oil. Hubei Agric Sci. 2012(13):46–49.
- Xu M, Lyu Y, Guo Z, Mo Z. 2020. Characterization and phylogenetic relationships analysis of the complete chloroplast genome of Carya ovata. Mitochondrial DNA Part B. 5(3):3087–3088.
- Yang X, Zhou T, Su X, Wang G, Zhang X, Guo Q, Cao F. 2020. Structural characterization and comparative analysis of the chloroplast genome of Ginkgo biloba and other gymnosperms. J Forest Res. 31: 1–14.
- Zhang JB, Li RQ, Xiang XG, Manchester SR, Lin L, Wang W, Wen J, Chen ZD. 2013. Integrated fossil and molecular data reveal the biogeographic diversification of the eastern Asian-eastern North American Disjunct Hickory Genus (Carya Nutt.). PLOS One. 8(7):e70449.