Abstract
Rhodiola kirilowii is a widely used Tibetan medicine. Here, we report the complete sequence of the chloroplast genome of R. kirilowii. The genome was 150,896 bp in length with 131 genes comprising 85 protein-coding genes, 37 tRNA genes, 8 rRNA genes and 1 pseudogene, with 20 of them occurring in double copies. Phylogenomic analysis suggested that R. kirilowii forms a clade with R. rosea, R. yunnanensis, R. fastigiata and R. crenulata in Rhodiola genus.
Rhodiola kirilowii is a herbaceous perennial belonging to the family Crassulaceae (Fu and Ohba Citation2001). Rhodiola kirilowii is an important Tibetan medicine, from which various distinct groups of chemical compounds (phenylethanol derivatives) have been isolated (Yang Citation1991; Grech-Baran et al. Citation2015). It was demonstrated that R. kirilowii could be effective in increasing immunity, stimulating nervous system, eliminating fatigue and preventing high-altitude sickness (Panossian et al. Citation2010). Here, we report the completed chloroplast genome of R. kirilowii and investigate its phylogenetic relationship within Crassulaceae, which may be conductive to a deeper understanding of this particular species.
Rhodiola kirilowii was collected from Baoku Forestry Farm (Qinghai, China;N 37°07′06″, E 101°34′42″). The voucher specimen (Chi202001) was stored in Qinghai-Tibetan Plateau Museum of Biology (HNWP). Total genomic DNA was obtained from silica-gel dried leaves of R. kirilowii by modified CTAB method (Doyle Citation1987). Sequencing was performed on the Illumina HiSeq2500 platform (San Diego, CA, USA). The chloroplast genome was assembled by SPAdes (Bankevich et al. Citation2012), annotated by CpGAVAS (Liu et al. Citation2012).
A typical quadripartite structure with a total length of 150,896 bp was found in the R. kirilowii chloroplast genome. The large single-copy region (LSC, 82,233 bp) and the small single-copy region (SSC, 16,992 bp) were separated by two inverted repeats (IR, 25,834 bp). Totally 131 genes were predicted in the genome, including 85 protein-coding genes, 37 tRNA genes, 8 rRNA genes and 1 pseudogene. 9 protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1, and ycf2), 4 rRNA genes (4.5S, 5S, 16S, and 23S rRNA) and 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) duplicated in the IR regions.
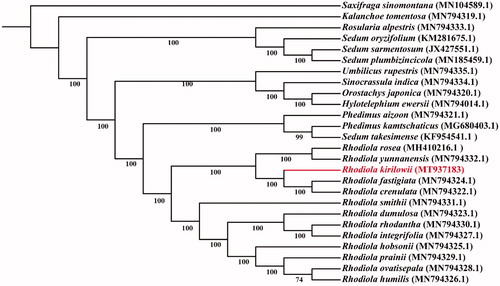
Phylogenetic analysis was performed among R. kirilowii and 24 species in Crassulaceae with Saxifraga sinomontana as the outgroup. The whole plastome sequences were used for the phylogenetic analysis. Phylogenetic analysis was conducted on the PhyloSuite platform (Zhang et al. Citation2020). All of the sequences were aligned using MAFFT. ModelFinder were used to select the best model for phylogenetic analyses. The phylogenetic trees were constructed using IQ-TREE with 1000 bootstraps. All the generated trees were modified by Interactive Tree Of Life (iTOL, http://itol.embl.de/) (). The phylogenetic tree showed that R. kirilowii forms a clade with R. rosea, R. yunnanensis, R. fastigiata and R.crenulata.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The chloroplast genome and raw sequencing data in this study are available in GenBank (https://www.ncbi.nlm.nih.gov/) under the accession numbers of MT937183 and SRR12991102.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu KT, Ohba H. 2001. Crassulaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 8. Beijing: Science Press. p. 251-268.
- Grech-Baran M, Sykłowska-Baranek K, Pietrosiuk A. 2015. Approaches of Rhodiola kirilowii and Rhodiola rosea field cultivation in Poland and their potential health benefits. Ann Agric Environ Med. 22(2):281–285.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13:715
- Panossian A, Wikman G, Sarris J. 2010. Rosenroot (Rhodiola rosea): traditional use, chemical composition, pharmacology and clinical efficacy. Phytomed. 17(7):481–493.
- Yang YC. 1991. Flora Tibetan Medicine. Xininng, China: Qinghai People's Publishing House.
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.