Abstract
The complete chloroplast genome of Geum macrophyllum is reported and characterized in this study. The chloroplast genome of G. macrophyllum was a circular form with a size of 155,940 bp in length. The genome presented a typical quadripartite structure composed of a pair of inverted repeats (IRa and IRb) of 26,152 bp separated by a large single copy (LSC) region of 85,307 bp and a small single copy (SSC) region of 18,329 bp. The genome contained a set of 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes, in which 17 were duplicated and 112 were unique. Phylogenetic analysis placed G. macrophyllum as sister to G. triflorum based on current sampling.
Geum macrophyllum Willd. belongs to the family Rosaceae Juss., subfamily Rosoideae (Juss.) Arn., and tribe Colurieae Rydb. This species is distributed in Eurasia and North America (Rohrer Citation2014). According to Plants of the World online maintained by the Royal Botanic Gardens, Kew, UK (http://powo.science.kew.org), its native range is from Kamchatka to North and Central Japan, as well as North America. G. macrophyllum has been used as traditional medicine (McCutcheon et al. Citation1994; Ellsworth et al. Citation2013) and its roots contain compounds which are effective against fungal diseases (McCutcheon et al. Citation1994). The complete chloroplast (cp) genome of G. macrophyllum reported herein provides a foundation for further studies on its taxonomy, evolution, and population genetics.
Voucher specimen for G. macrophyllum (J. Wen 10,445) was collected from Quebec, Canada, and deposited in the United States National Herbarium (US). Total genomic DNA was isolated from silica gel-dried leaves using the method of Doyle and Doyle (Citation1987). The library with insert size of 300 bp fragments was constructed using the KAPA Hyper Prep Kit for Illumina® and then sequenced using the Illumina HiSeq platform in Novogene (Davis, CA). After sequencing, adapters were removed by the trimming software Trimmomatic version 0.33 (Bolger et al. Citation2014). The number and quality of raw paired‐end reads were evaluated by using the FastQC (Andrews Citation2018). The raw paired‐end reads were used to assemble the cp genome in NOVOPlasty version 3.8.2 (Dierckxsens et al. Citation2017), with ribulose-1, 5-bisphosphate carboxylase/oxygenase (rbcL) gene from G. rupestre (T. T. Yü et C. L. Li) Smedmark (GenBank accession no. NC_037392) as the seed sequence. Chloroplast genome annotation of G. macrophyllum was first performed using GeSeq (Tillich et al. Citation2017), with complete chloroplast genome of G. rupestre (NC_037392), Fragaria chiloensis (L.) Mill. (NC_019601) and Farinopsis salesoviana (Steph.) Chrtek et Soják (MT017928) as reference sequences. Draft annotation generated by GeSeq was then imported into Geneious Prime (Kearse et al. Citation2012) for further manual adjustment. Where necessary, gene boundaries were corrected to match the start and stop codons and intron/exon boundaries. The annotated complete cp genome of G. macrophyllum (accession no. MT774132) was submitted to GenBank. The complete cp genome of G. macrophyllum was a circular DNA molecule with a size of 155,940 bp in length. The genome had a typical quadripartite structure composed of two copies of inverted repeats (IRa and IRb: 26,152) separated by a large single-copy region (LSC: 85,307 bp) and a small single-copy region (SSC: 18,329 bp). The overall GC content was 36.6%, and that of the LSC, SSC, and each IR were 34.3%, 30.6%, and 42.6%, respectively. The cp genome encoded a set of 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes, in which 112 were unique and 17 were duplicated. The 17 duplicated genes in IR regions contained six protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23).
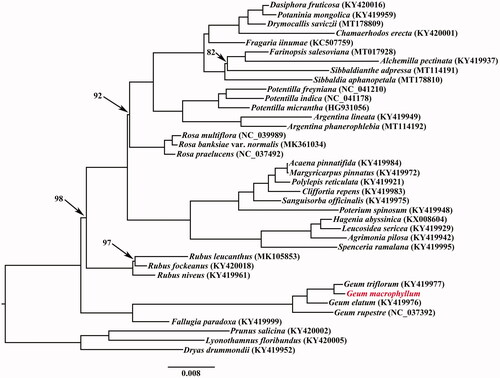
In order to investigate the phylogenetic position of G. macrophyllum, the cp genome sequences of 38 Rosaceae taxa were aligned with MAFFT version 7.450 (Katoh and Standley Citation2013) and then trimmed properly by trimAL version 1.4 (Capella-Gutiérrez et al. Citation2009). A maximum likelihood (ML) phylogenetic analysis was conducted using RAxML version 8 (Stamatakis Citation2014) following Zhang et al. (Citation2020). The phylogenetic tree placed G. macrophyllum as sister to G. triflorum based on current sampling ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MT774132. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA670790, SRP288241, and SAMN16520620, respectively.
Additional information
Funding
References
- Andrews S. 2018. Fast QC: a quality control tool for high throughput sequence data. [accessed 2020 March 8]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Ellsworth KT, Clark TN, Gray CA, Johnson JA. 2013. Isolation and bioassay screening of medicinal plant endophytes from eastern Canada. Can J Microbiol. 59(11):761–765.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- McCutcheon AR, Ellis SM, Hancock REW, Towers GHN. 1994. Antifungal screening of medicinal plants of British Columbian native peoples. J Ethnopharmacol. 44(3):157–169.
- Rohrer JR. 2014. Geum L. 1993+. Flora of North America North of Mexico. 19+ vols. Vol. 9. Oxford: Oxford University Press; p. 58–70.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhang XH, Khasbagan , Li QQ. 2020. The complete chloroplast genome of Sibbaldia aphanopetala (Rosaceae: Potentilleae). Mitochondrial DNA Part B. 5(3):2026–2027.