Abstract
In this study, we report the first complete plastome sequence of Vitex rotundifolia (Lamiaceae) (MT937186). In addition, the plastome sequences of Phryma leptostachya subsp. asiatica (Phrymaceae) (153,324 bp; MT948145) and Mazus pumilus (Mazaceae) (152,847 bp; MT937187) are also included. The gene orders and structures of the three plastomes are collinear with those of the typical plastome of angiosperm. The plastome size of V. rotundifolia is 154,370 bp in length and consists of a large single-copy region of 85,079 bp and a small single-copy region of 17,917 bp, which are separated by a pair of 25,687 bp-long inverted repeat regions. In addition, the plastome sizes of P. leptostachya subsp. asiatica and M. pumilus are 153,324 bp and 152,847 bp, respectively. The three plastomes contain 113 genes, including 79 protein-coding, 30 tRNA, and four rRNA genes. Sixteen genes contain one intron and two genes have two introns. A total of 41 simple sequence repeat loci was identified in the V. rotundifolia plastome. Phylogenetic analysis shows that Viticoideae is a sister group of the last of Lamiaceae except Nepetoideae. The Mazaceae are a sister group of Lamiaceae, while Phrymaceae form a sister group to the Paulowniaceae-Orobanchaceae clade.
Vitex rotundifolia L.f. is a small shrub and native to seashores throughout the pacific side of Northeast Asia. It belongs to the family Lamiaceae in the order Lamiales (APG IV Citation2016). Lamiaceae consist of 12 subfamilies, 241 genera, and approximately 7530 species (Christenhusz and Byng Citation2016). The genus Vitex belongs to the subfamily Viticoideae of Lamiaceae. Phryma leptostachya L. subsp. asiatica (H. Hara) Kitamura (Phrymaceae) is a medium size annual herb and native to Northeastern America and Asia. It belongs to the family Phrymaceae in the order Lamiales (APG IV Citation2016). Mazus pumilus (Burm.f.) Steenis (Mazaceae) is a small annual herb in the open damp locations and native to South and East Asia. It belongs to the family Mazaceae in the order Lamiales (APG IV Citation2016). Phryma leptostachya subsp. asiatica (Phrymaceae) and Mazus pumilus (Mazaceae) belong to the sister families of Lamiaceae.
Samples of V. rotundifolia, P. leptostachya subsp. asiatica, and M. pumilus were collected in South Korea and their GPS co-ordinations are N33°18′12.6″-E126°48′33.6″, N35°16′35.7″-E127°28′35.0″, and N37°19′18.5″-E127°15′44.6″, respectively. The fresh leaves were ground into powder in liquid nitrogen and total DNAs were extracted using the G-spinTM IIp for Plant Genomic DNA Extraction Kit (iNtRON Biotechnology, Seongnam-si, South Korea). The voucher specimens were deposited in the Korea University Herbarium (KUS acc. nos. 2008-0463, 2008-0813, and 2008-1240) and their genomic DNAs were deposited in the Plant DNA Bank in Korea (PDBK acc. no. 2008-0463, 2008-0813, and 2008-1240). NGS sequencings were performed using an Illumina MiSeq platform (Illumina Inc., San Diego, CA). Approximately, 100 ng of extracted DNA were used for library construction and raw sequence reads were generated using Illumina MiSeq using reagent kit v3 (600-cycles) (Illumina, Inc., San Diego, CA). The average length of raw reads is 301 bp. De novo assemblies and annotations of plastome were performed using the Geneious 11.1.5 (Biomatters Ltd., Auckland, New Zealand; Kearse et al. Citation2012), National Center for Biotechnology Information (NCBI) BLAST, and tRNAscan-SE programs (Lowe and Eddy Citation1997). The plastome of Sesamum indicum (NC016433) was used as a reference genome for annotation (Yi and Kim Citation2012). The average plastome coverage of V. rotundifolia, P. leptostachya subsp. asiatica., and M. pumilus is 2003×, 505×, and 778×, respectively. The simple sequence repeats (SSRs) were detected with the Phobos v. 3.3.12 program (Leese et al. Citation2008) in the Geneious 11.1.5. For the phylogenetic analysis, we selected and downloaded 27 related complete plastome sequences based on the APG IV system (APG IV Citation2016) from the NCBI database.
The gene orders and structures of the three plastomes are collinear to those of typical angiosperms (Shinozaki et al. Citation1986; Kim and Lee Citation2004; Yi and Kim Citation2012). The complete plastome of V. rotundifolia is 154,370 bp in length, and consists of a large single-copy (LSC) region of 85,079 bp and a small single-copy (SSC) region of 17,917 bp, which are separated by two inverted repeats (IRs) of 25,687 bp. The plastome comprises 113 unique genes (79 protein-coding genes, 30 tRNA genes, and four rRNA genes). Six protein-coding, seven tRNA, and four rRNA genes are duplicated in the IR regions. The average A-T content of the plastome is 61.7%, whereas that in the LSC, SSC, and IR regions is 63.6%, 67.3%, and 56.7%, respectively. Sixteen genes contain one intron and two genes, ycf3 and clpP, have two introns. A total of 41 SSR loci are distributed throughout the plastome. Among these, 35, 5, and 1 are mono-SSR, di-SSR, and tri-SSR loci, respectively.
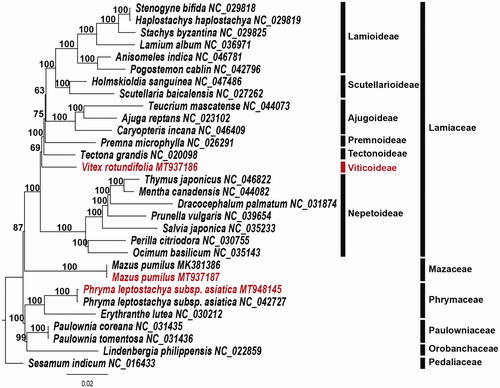
The complete plastomes of P. leptostachya subsp. asiatica (153,324 bp; MT948145) and M. pumilus (152,847 bp; MT937187) were also fully assembled and annotated. The plastome size of P. leptostachya subsp. asiatica is 157 bp longer than the previously reported same species (Xia et al. Citation2019). In contrast, the plastome size of M. pumilus was 187 bp shorter than the previously reported same species (Xia et al. Citation2019). These differences probably reflect the geographic differences between Korean and Chinese populations. To validate the phylogenetic relationships of three species, 30 whole plastome sequences including all noncoding regions were aligned as a single data matrix (178,589 bp in length) using the MAFFT v. 7.017 in Geneious v. 11.1.5 (Biomatters Ltd., Auckland, New Zealand; Kearse Citation2012). Then, a maximum-likelihood (ML) tree was reconstructed by RAxML v.8.2.12 in CIPRES webserver (Stamatakis Citation2014) using the GTR + G + I model with 1000 bootstrap replicates. The resulting tree shows that V. rotundifolia (Viticoideae) is a sister group of the last of Lamiaceae except Nepetoideae (). In previous studies, Viticoideae show a close relationship to Symphorematoideae (Li et al. Citation2016; Zhao et al. Citation2020). Mazaceae (M. pumilus) were the sister family of Lamiaceae, while Phrymaceae (P. leptostachya subsp. asiatica) form a sister group to the Paulowniaceae-Orobanchaceae clade. The complete plastome sequence in this report will provide a useful resource for the phylogenetic and evolutionary studies of Lamiales.
Disclosure statement
The authors report no conflicts of interest, and are independently responsible for the content and writing of the paper.
Data availability statement
The data that support the finding of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT937186 for Vitex rotundifolia, MT948145 for Phryma leptostachya subsp. asiatica, and MT937187 for Mazus pumilus.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Christenhusz MJ, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa. 261(3):201–217.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim K-J, Lee H-L. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11(4):247–261.
- Leese F, Mayer C, Held C. 2008. Isolation of microsatellites from unknown genomes using known genomes as enrichment templates. Limnol Oceanogr Methods. 6(9):412–426.
- Li B, Cantino PD, Olmstead RG, Bramley GLC, Xiang C-L, Ma Z-H, Tan Y-H, Zhang D-X. 2016. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci Rep. 6:34343.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5(9):2043–2049.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xia Z, Wen J, Gao Z. 2019. Does the enigmatic Wightia belong to Paulowniaceae (Lamiales)? Front Plant Sci. 10:528.
- Yi D-K, Kim K-J. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLOS One. 7(5):e35872.
- Zhao F, Li B, Drew BT, Chen Y-P, Wang Q, Yu WB, Liu ED, Salmaki Y, Peng H, Xiang C-L. 2020. Leveraging plastomes for comparative analysis and phylogenomic inference within Scutellarioideae (Lamiaceae). PLOS One. 15(5):e0232602.