Abstract
Canavalia gladiata is a member of genus Canavalia, which includes 60 species around world. And the species in genus of Canavalia have similar character so we use complete chloroplast genome to identify C. gladiata and other species. Here, we assembled and analyzed the complete chloroplast (cp) genome of C. gladiata. The cp genome of C. gladiata is 157,923 bp, with a total GC content of 34.6%. The complete cp genome has a typical quadripartite structure, including a large single-copy (LSC) region of 77,660 bp, a small single-copy (SSC) region of 18,945 bp and two inverted repeat (IRS) regions of 30,659 bp. The complete cp genome contains 136 genes, including 91 protein-coding genes, 37tRNA and 8 rRNA genes. Phylogenetic analysis result showed that C. gladiata is closely related to C. rosea.
Canavalia gladiata is a member of genus Canavalia in subfamily Papilionoideae and the genus Canavalia includes about 60 species around the world (Sauer Citation1964; Aymard and Cuello Citation1991). Canavalia gladiata is well known as originated in Asian, and distributed around Asia west Indian, Africa, South America and Australia (Herklots Citation1972; Kay Citation1979). Canavalia gladiata was used as food, Chinese herbal medicine and sauce of phytochemical and pharmaceutical products (Nanda et al. Citation1993; Eknayake et al. Citation1999; Arun et al. Citation2003). Nowadays, he chloroplast genome plays an important role in species identification by its characters such as uniparental inheritance, conserved sequence composition in coding regions and numerous variable sites (Bi et al. Citation2018). However, the published complete chloroplast genome were no more than 3 species in this genus and C. gladiata was not included. In this study, we sequenced and identified the complete chloroplast genome of C. gladiata.
We selected the sample in Yibin Botanical Garden (27°50′N, 103°36′E) on 1 August 2020. The voucher specimen (CG20208264ZJ) was deposited in Laboratory of Molecular Biology in YiBin Vocational and Technical College. We used the fresh leaves to extract DNA with the modified CTAB method (Doyle and Doyle Citation1987) and constructed the libraries with an average length of 350 bp using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA), then the libraries were sequenced on Illumina Novaseq 6000 platform, 2.81 Gb clean data was assembled by SPAdes v.3.11.0 software (Bankevich et al. Citation2012) using the chloroplast genome of Canavalia rosea (Accession number: LC554221) as the reference. Finally, the assembled complete cp genome was annotated by Plann software (Huang and Cronk Citation2015), and submitted to GenBank under the accession number of MT922037.
The total length of complete cp genome of C. gladiata is 157,923 bp, with a total GC content of 34.6%. The complete cp genome has a typical quadripartite structure, including a large single copy (LSC) region of 77,660 bp, a small single copy (SSC) region of 18,945 bp and two inverted repeat (IRS) regions of 30,659 bp. The complete cp genome contains 136 genes, including 91 protein coding genes, 37tRNA and. 8 rRNA genes. trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, ndha genes contained an intron, and clpP and ycf3 contained 2 introns.
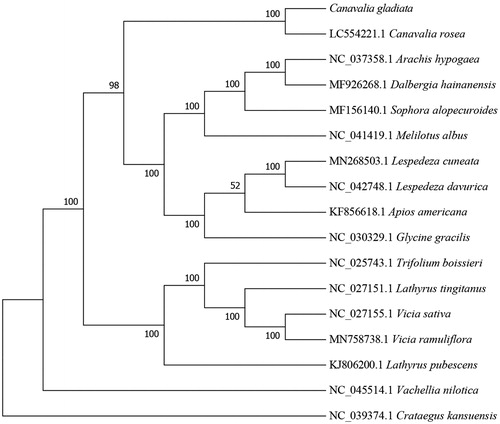
To determine the phylogenetic position of C. gladiata, the complete chloroplast genome of C. gladiata was aligned with other 15 species in subfamily Papilionoideae from GenBank using Mafft-7.037 (Katoh and Standley Citation2013). Subsequently, the phylogenetic tree was constructed by IQTREE v1.6 (Nguyen et al. Citation2015; Hoang et al. Citation2018) with 1000 bootstraps replicates using Best-fit model. By using Crataegus kansuensis(NC_039374.1) as out group we got the result that C. gladiata was closely related to C. rosea (LC554221.1) (). These information will provide useful genomic resources for the development of molecular markers and accurate identification of C. gladiata.
Figure 1. Maximum-likelihood phylogenetic tree for Canavalia gladiata based on 17 complete chloroplast genomes(Canavalia rosea, Vachellia nilotica, Arachis hypogaea, Dalbergia hainanensis, Vicia sativa, Lathyrus tingitanus, Vicia ramuliflora, Lespedeza cuneata, Sophora alopecuroides, Glycine gracilis, Lathyrus pubescens, Trifolium boissieri, Apios americana, Lespedeza davurica, Melilotus albus, Canavalia gladiata and Crataegus kansuensis). The Genbank accession numbers are on the diagram.
Disclosure statement
No potential conflict of interest was reported by the author.
Data availability statement
The assembled complete chloroplast genome sequence of Canavalia gladiata has been submitted to GenBank under the accession number: MT922037 (https://www.ncbi.nlm.nih.gov/nuccore/MT922037).
References
- Arun AB, Sridhar KR, Raviraja NS, Schmidt E, Jung K. 2003. Nutritional and antinutritional components of Canavalia spp. seeds from the West Coast sand dunes of India. Plant Foods Hum Nutr. 58(3):1–13.
- Aymard, C, G , and A. N. Cuello . "Catálogo y adicciones a las especies neotropicales del género canavalia (Leguminosae - papilionoidea - Phaseoleae - diocleinae)." Fonaiap (1991).
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bi Y, Zhang MF, Xue J, Dong R, Du YP, Zhang XH. 2018. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Front Plant Sci. 8:1184.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Eknayeke S, Jansz ER, Nair BM. 1999. Proximate composition of mineral and amino acid content of mature Canavalia gladiata seeds. Food Chem. 66:115–119.
- Herklots GAC. 1972. Beans and peas. Vegetables in Southeast Asia. London: George Ellen and Unwind Ltd.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kay ED. 1979. Food legumes. TPI crops and product Digest No. 3. London: Tropical Products Institute; p. 435.
- Nanda IP, Pande RK, Kar PK. 1993. Food value of Canavalia gladiata seeds. Acta Bot Indica. 25:144–145.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sauer J. 1964. Revision of Canavalia. Brittonia. 16(2):106–181.
