Abstract
Nardostachys jatamans is an endemic herb in China, distributes mainly in Southeast Gansu, South Qinghai and West Sichuan of Qinghai-Tibet Plateau. In this study, the complete chloroplast genome (a typical quadripartite structure) sequence of N. jatamans was reported. The length of the DNA molecule was 155,268 bp with a large single-copy region (LSC: 87,263 bp), small single-copy region (SSC: 17,327 bp) and inverted repeats (IRa and IRb: 25,339 bp). The overall GC content was 38.56%. It has a total of 129 genes, containing 83 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The phylogenetic analysis has shown that N. jatamans is sister to Valeriana offcinalis. The chloroplast genome provides the basis for development and utilization of N. jatamans in future.
Nardostachys jatamans, locates in Southeast Gansu, South Qinghai and West Sichuan of Qinghai-Tibet Plateau in China, contains multiple chemical compositions including alkaloids, minor triterpenes, steroids, lignans, flavonoids, coumarins, lignan and terpenoids, etc. (Paek and Lim Citation2014), is an endemic traditional medicinal herb for its biological activities (anti-depressant, anti-arrhythmia, anti-oxidation, anti-tumor and anti-inflammation, etc.) (Wu et al. Citation2011; Liu et al. Citation2013; Zhang et al. Citation2015; Wu et al. Citation2018).
Fresh leaves of N. jatamans was collected from the Autonomous Prefecture of Aba Tibet and Qiang Race of Sichuan Province and stored in the refrigerator at −80 °C. Voucher specimen (No. GS-AB-01) was deposited in the herbarium of Guizhou Education University. Total genomic DNA was isolated by using a modified CTAB method (Doyle Citation1987). The library with insert size of 300 bp fragments was sequenced by Illumina Hiseq 2500 Sequencing System (Illumina, Hayward, CA, USA) in Genepioneer Biotechnologies Co. Ltd, Nanjing, China and the lib The NOVOPlasty software was used to assemble the raw reads (Dierckxsens et al. Citation2017). CLC Genomics Workbench v8.0 was used to filter out the low-quality sequences and the chloroplast genome was reconstructed by MITObim v1.8 (Hahn et al. Citation2013). The complete chloroplast genome of N. jatamans was annotated in Geneious Prime (Kearse et al. Citation2012) and aligning with relatively related species. The data of complete chloroplast genome sequence of N. jatamans were submitted to GenBank (MW149527). The complete cp genome of N. jatamans was 155,268 bp in length. There was a circular DNA molecule with typical quadripartite structure (IRa and IRb: 87,263 bp, LSC: 87,263 bp and SSC: 17,327 bp). The overall GC content was 38.56%. The GC content was 36.77%, 33.3% and 43.42% in the regions of LSC, SSC and IR, respectively. GC content of IRs region is the highest. There were 22 genes have only one intron, and three genes have two introns. There were 129 genes, containing 83 protein-coding genes, 38 tRNA genes, and eight rRNA genes in the complete chloroplast genome sequence of N. jatamans.
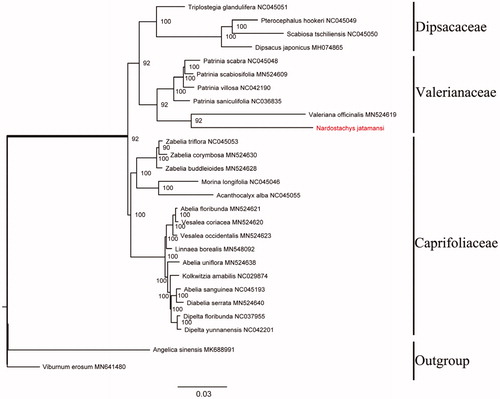
To investigate the phylogeny of N. jatamans, 26 taxa were included (15 species from Caprifoliaceae, 5 species from Valerianaceae, 4 species from Dipsacaceae and 2 species as outgroups). All sequences were aligned in MAFFT (Katoh and Standley Citation2013) and manually adjusted by using trimAL version 1.4 (Capella-Gutiérrez et al. Citation2009). RAXML version 8 (Stamatakis Citation2006) was used to infer maximum likelihood (ML) tree chosen GTRGAMMA model with 1000 rapid bootstrap. The phylogenetic tree has shown that N. jatamans and Valeriana offcinalis were fallen into a branch with robust bootstrap support values in Valerianaceae ().
Figure 1. Maximum-likelihood phylogenetic tree based on 27 complete chloroplast genomes. The ML bootstrap support value is on each branch. The Chloroplast genomic accession numbers used in this phylogeny analysis: Patrinia scabra (NC045048), Patrinia scabiosifolia (MN524609), Patrinia villosa (NC042190), Patrinia saniculifolia (NC036835), Valeriana officinalis (MN524619), Acanthocalyx alba (NC045055), Kolkwitzia amabilis (NC029874), Diabelia serrata (MN524640), Abelia floribunda (MN524621), Abelia sanguinea (NC045193), Abelia uniflora (MN524638), Pterocephalus hookeri (NC045049), Dipelta yunnanensis (NC042201), Morina longifolia (NC045046), Dipelta floribunda (NC037955), Vesalea occidentalis (MN524623), Vesalea coriacea (MN524620), Zabelia triflora (NC045053), Zabelia corymbosa (MN524630), Zabelia buddleioides (MN524628), Kolkwitzia amabilis (NC029874), Linnaea borealis (MN548092), Triplostegia glandulifera (NC045051), Dipsacus japonicus (MH074865), Scabiosa tschiliensis (NC045050), Viburnum erosum (MN641480), and Angelica sinensis (MK688991).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW149527.
Additional information
Funding
References
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu ML, Duan YH, Hou YL, Li C, Gao H, Dai Y, Yao XS. 2013. Nardoaristolones A and B, two terpenoids with unusual skeletons from Nardostachys chinensis Batal. Org Lett. 15(5):1000–1003.
- Paek JH, Lim SS. 2014. Preparative isolation of aldose reductase inhibitory compounds from Nardostachys chinensis by elution-extrusion counter-current chromatography. Arch Pharm Res. 37(10):1271–1279.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wu J, Shi J, Liu Y, Yan X, Liu Y. 2011. Herbal verification of Nardostachys jatamans. J Chin Med Mater. 034:1459–1461.
- Wu PQ, Yu YF, Zhao Y, Yu CX, Zhi DJ, Qi FM, Fei DQ, Zhang ZX. 2018. Four novel sesquiterpenoids with their anti-Alzheimer's disease activity from Nardostachys chinensis. Org Biomol Chem. 16(46):9038–9045.
- Zhang JB, Liu ML, Li C, Zhang Y, Dai Y, Yao XS. 2015. Nardosinane-type sesquiterpenoids of Nardostachys chinensis Batal. Fitoterapia. 100:195–200.
