Abstract
Gardenia jasminoides Ellis is a traditional aromatic and medicinal plant in China. Here, the complete chloroplast genome of a wild-type gardenia adapted to island climate was assembled. The assembled genome was 155,247 bp in length, with four typical regions, i.e., a large single-copy (LSC) region (85,414 bp), a small single-copy (SSC) region (18,235 bp) and two inverted repeats (IRs) regions (25,799 bp each). In total, 138 genes were predicted, including 90 protein-coding genes, 40 tRNA genes and eight rRNA genes. The overall GC content of the chloroplast genome was 37.5%. The chloroplast genome would provide more information for the phylogeography and phylogeny study of G. jasminoides.
Gardenia jasminoides Ellis, belonging to the family Rubiaceae, is a traditional aromatic and medicinal plant. The main officinal part of G. jasminoides is its dried ripe fruit. Chemical composition analysis finds that there are multiple secondary metabolites in the fruit of G. jasminoides, like iridoid glycosides, crocin and crocetin, which have broad spectrum anti-inflammatory properties and even anti-cancer effect (Qin et al. Citation2013; Chen et al. Citation2015, Citation2017). All these make G. jasminoides an important resource plant, and relative studies are arising. Zhao and Zhou (Citation2020) reported the complete chloroplast genome of wild-type gardenia based on the material collected from Quanzhou, Fujian province, China. However, G. jasminoides is widely distributed in the area south of the Yangtze River, which is adapted to divergent habitats and may form different genotypes. In this study, the complete chloroplast genome of another wild-type gardenia adapted to island climate was assembled, which could provide more information for the phylogeography and phylogeny study of this species.
The material used in this study was collected from Nanji islands, Wenzhou, Zhejiang province of China (121°5′44.75″ E, 27°27′21.58″ N). The voucher specimen was kept in the herbarium of Nanjing Forestry University (accession number: NF2019102301). Fresh leaves of G. jasminoides were harvested and frozen in liquid nitrogen, then transferred to −80 °C refrigerator, immediately. Genetic DNA was extracted from these leaves for sequencing library construction. The library was then paired-end sequenced on the Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA) by Genepioneer Biotechnologies Co., Ltd. (Nanjing, China) with the standard Illumina re-sequencing protocols. In total, 6 Gb clean reads were obtained. After reads quality filtration, SPAdes 3.11.0 (Bankevich et al. Citation2012) and SSPACE (Boetzer et al. Citation2011) were alternately used for data assembly. Then, GapFiller v1.11 (Boetzer and Pirovano Citation2012) was further applied for gap filling. The complete sequence was primarily annotated by PGA (Qu et al. Citation2019) combined with GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html). Finally, IRscope (https://irscope.shinyapps.io/irapp/), an online tool was used to identify the borders of IR, LSC and SSC regions. The chloroplast genome of G. jasminoides assembled in this study was 1,55,247 bp in length with four typical regions. The lengths for LSC, SSC and IRs regions were 85,414 bp, 18,235 bp and 25,799 bp, respectively. So, it is longer than that reported by Zhao and Zhou (Citation2020) for two insertion mutations at non-coding regions. The whole LSC region of G. jasminoides in Zhao and Zhou’s (Citation2020) study was inverted, but inversion mutation was not detected in this work. Excluding these mutations, the identity between the two genomes is 99.99%. Finally, 138 genes were predicted in our study, including 90 protein-coding genes (81 species), 40 tRNA genes (32 species) and 8 rRNA genes (4 species). The overall GC content of the chloroplast genome was 37.5%.
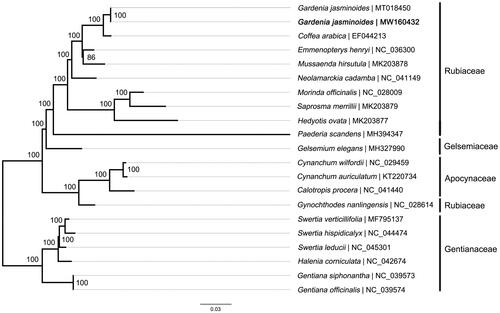
Maximum likelihood (ML) tree was constructed to reveal the phylogenetic relationship of G. jasminoides with other species in Gentianales. Genome-wide alignment was conducted by MAFFT 7.158 (Katoh and Standley Citation2013). Then phylogenetic inference was performed in RAxML-VI-HPC (Stamatakis Citation2006) software under the GTR-gamma model. To assess the confidence of each internal node, rapid bootstrap method was applied with 1000 replications. The result showed that phylogenetic positions of all the taxa were successfully resolved (). Gardenia jasninoides was placed in Rubiaceae clade and classed together with Coffea arabica, which indicated that Gardenia is more closely related to Coffea than other studied genera.
Figure 1. Maximum-likelihood tree based on the sequences of 21 chloroplast genomes from Gentianales. Numbers at tree nodes represent bootstrap values for 1000 replications. Number after ‘|’ shows the accession number in GenBank for each accession. The position of the wild-type Gardenia jasminoides reported in this study is marked in bold.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The raw sequence data supporting this study are deposited in the National Center for Biotechnology Information Short Read Archive under BioProject ID PRJNA678106 (accession number SRP292439). The assembled genome and its annotation are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW160432.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 27(4):578–579.
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13(6):R56.
- Chen S, Zhao S, Wang X, Zhang L, Jiang E, Gu Y, Shangguan AJ, Zhao H, Lv T, Yu Z. 2015. Crocin inhibits cell proliferation and enhances cisplatin and pemetrexed chemosensitivity in lung cancer cells. Transl Lung Cancer Res. 4(6):775–783.
- Chen SC, Zhao X, Yi RK, Qian J, Shi YH, Wang R. 2017. Anticancer effects of Gardenia jasminoides in HepG2 human hepatoma cells. Biomed Res. 28:716–726.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Qin FM, Meng LJ, Zou HL, Zhou GX. 2013. Three new iridoid glycosides from the fruit of Gardenia jasminoides var. radicans. Chem Pharm Bull. 61(10):1071–1074.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Zhao K, Zhou Y. 2020. The chloroplast genome of Gardenia jasminoides and related phylogenetic analysis (Rubiaceae). Mitochondrial DNA Part B. 5(2):1743–1745.
