Abstract
Sargassum hemiphyllum (Turner) C. Agardh is an important brown macroalga. In this study, we presented the complete chloroplast genome of its variety S. hemiphyllum var. chinense using genome skimming approach. Circular mapping revealed its sequence length was 124,319 bp, with a large single-copy region (LSC, 73,505 bp) and a small single copy region (SSC, 39,922 bp) separated by a pair of inverted repeats (IRs, 5446 bp). Its chloroplast genome contained 173 genes, including 139 protein-coding, 6 rRNA, and 28 tRNA genes. The phylogenetic analysis indicated that S. hemiphyllum var. chinense was closely related with S. confusum.
Sargassum hemiphyllum (Turner) C. Agardh, a kind of brown macroalga that belongs to the Sargassaceae, distributed from Japanese and Korean coasts to the southern Chinese and northern Vietnam coasts (Cheang et al. Citation2010). As one of the important seaweeds for mariculture, S. hemiphyllum is widely applied as food, polysaccharide source, and herbal medicine in China (Cheang et al. Citation2008). According to previous research, it was divided into two varieties: S. hemiphyllum var. chinense from China and Vietnam, and S. hemiphyllum var. hemiphyllum from Japan and Korea (Ajisaka et al. Citation1997; Cheang et al. Citation2008). Despite the importance of the species, there has been no genomic studies on S. hemiphyllum except the study of its mitochondrial genome (Liu et al. Citation2016). In this study, we obtained and characterized the complete chloroplast genome of S. hemiphyllum var. chinense and clarified the phylogenomic relationship with other species in the Phaeophyceae.
One S. hemiphyllum var. chinense individual was collected from Shenzhen, Guangdong Province of China (N 22°32′52.56″, E 114°34′27.67″), and its specimen was stored at Zhejiang Mariculture Research Institute (Dongtou Base) with an accession number Wu202001. Total DNA was extracted using the modified CTAB method (Doyle and Doyle Citation1990). Paired-end reads were sequenced on the NovoSeqPE150 platform (Novogene Co., LTD). The draft complete chloroplast genomes were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017). The chloroplast genome of Sargassum horneri (GenBank accession number: KP881334) was used as the reference sequence. Then, paired-end reads were mapped to the draft genome using CLC Genomics Workbench 11 and yielded the complete chloroplast genome sequence of S. hemiphyllum var. chinense. Gene annotation was conducted via the online program Dual Organellar Genome Annotator (DOGMA; Wyman et al. Citation2004) using the same method described by Zhang et al. (Citation2019).
The complete chloroplast genome of S. hemiphyllum var. chinense (GenBank accession MT800998) is a circular DNA molecule measuring 124,319 bp in length. It comprised a pair of inverted repeat regions (IRs with 5446 bp) divided by two single-copy regions (LSC with 73,505 bp and SSC with 39,922 bp). The nucleotide composition was 34.66% A (43,090 bp), 14.89% C (18,516 bp), 15.68% G (19,493 bp), 34.77% T (43,220 bp). The cp genome encoded a total of 173 genes, including 139 protein-coding, 6 rRNA, and 28 tRNA genes.
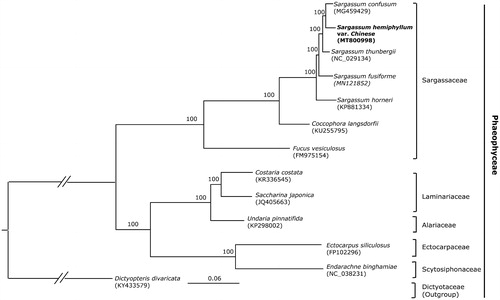
All 6 Sargassaceae species with complete chloroplast genome available in the NCBI were selected to reconstruct the phylogenomic tree. Then, maximum likelihood (ML) analyses were implemented on a data set that included 126 protein-coding genes for 12 taxa in Phaeophyceae using RAxML-HPC BlackBox (8.2.12) on CIPRES (http://www.phylo.org) under the GTR model. The phylogenetic tree showed a good resolution of the species of Sargassaceae and other families in Phaeophyceae, with full support at all the nodes. The phylogenomic result showed that S. hemiphyllum var. chinense is closely related with S. confusum ().
Disclosure statement
The authors are really grateful to the opened raw genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in Genbank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT800998. Sequence data of Sargassum hemiphyllum var. chinense from this article can be found in the NCBI database under BioProject codes PRJNA660488, Sequence Read Archive (SRA) ID SRR12558898.
Additional information
Funding
References
- Ajisaka T, Nang HQ, Dinh NH, Lu BR, Chiang YM, Yoshida T. 1997. Sargassum hemiphyllum (Turner) C. Agardh var. chinense J. Agardh from Vietnam, the Chinese mainland, and Taiwan (Fucales, Phaeophyta). In: Abbott IA, editor. Taxonomy of economic seaweeds with reference to some pacific species. Vol. 6. La Jolla (CA): California Sea Grant College Program, University of California.
- Cheang CC, Chu KH, Ang POA. 2010. Phylogeography of the marine macroalga Sargassum hemiphyllum (Phaeophyceae, Heterokontophyta) in northwestern Pacific. Mol Ecol. 19(14):2933–2948.
- Cheang CC, Chu KH, Ang PO. 2008. Morphological and genetic variation in the populations of Sargassum hemiphyllum (Phaeophyceae) in The Northwestern Pacific. J Phycol. 44(4):855–865.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Liu F, Pang SJ, Chen WZ. 2016. Complete mitochondrial genome of the brown alga Sargassum hemiphyllum (Sargassaceae, Phaeophyceae): comparative analyses. Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1468–1470.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang YH, Lee JK, Liu XL, Sun SZ. 2019. The first complete chloroplast genome of Hylomecon japonica and its phylogenetic position within Papaveraceae. Mitochondrial DNA Part B. 4(2):2349–2350.