Abstract
Scurrula chingii (W.C. Cheng) H.S. Kiu is a stem hemiparasite of the genus Scurrula in the family Loranthaceae distributed in southwest China and northern Vietnam. Here, we report and characterize the complete plastid genome sequence of S. chingii to provide genomic resources useful for the phylogenetic studies of Santalales. The plastome of S. chingii is 122,764 bp in length, consisted of a large single-copy region (70,726 bp), a small single-copy region (6,091 bp), and a pair of inverted repeat regions (22,974 bp). The GC content of the whole plastome is 37.2%. It contains 109 genes, including 69 CDS (protein-coding genes), eight rRNAs, and 32 tRNAs. The alignment of 14 species complete chloroplast genomes of Loranthaceae was implemented and a phylogenetic tree was constructed using maximum-likelihood (ML) method, which revealed that S. chingii clustered with Scurrula parasitica and Taxillus thibetensis as a monophyletic group.
Scurrula chingii (W.C. Cheng) H.S. Kiu is a stem hemiparasite distributed in southwest China (Guangxi and Yunnan provinces) and northern Vietnam at varied elevations from foothills to mountain ranges (http://www.efloras.org/). Although these plants conduct photosynthesis, they obtain a part of water and nutrients by parasitism, which is considered to be a pest as its infection intensity is high (Press and Phoenix Citation2005). As a generalist mistletoe, S. chingii can infect more than 38 host species (29 genera, 21 families) such as Camellia oleifera (Theaceae), Vernicia montana (Euphorbiaceae), and Ficus hispida (Moraceae) in Xishuangbanna, Southwest China (Wang and Zhang Citation2017). S. chingii can be usually found in altitudes between 490 and 1745 m a.s.l. in open forests and plantations and is less common in humid forests in Xishuangbanna, and dependent on frugivores birds for their pollination and seed dispersal (Wang and Zhang Citation2017). Here, we report the complete plastome of S. chingii based on Illumina paired-end sequencing data, reconstruct a phylogenetic tree and explore the evolutionary relationship of Loranthaceae, which will contribute to understanding the basic biological characteristics and role of mistletoe in the ecosystem further.
Fresh leaves of S. chingii were collected from Jinghong, Yunnan, Southwest China (Long. 101.097227 E, Lat. 22.428739 N, 860 m). The voucher specimen (accession number: LYJ-21) was deposited in the laboratory of the Research Group ‘Ecology and Evolution of Plant–Animal Interaction’ at Xishuangbanna Tropical Botanical Garden. Genomic DNA was extracted from the silica gel dried leaves using the modified cetyltrimethylammonium bromide (mCTAB) method (Li et al. Citation2013). Then, it was sheared into fragments to build Illumina libraries. The NGS (next generation sequencing) library was sequenced on Illumina HiSeq 2500 platform using 150 bp paired-end strategy. Meanwhile, the raw sequence data were uploaded on the NCBI (SRA accession number: PRJNA666529). The plastome of S. chingii was assembled into circular form from the raw reads using Getorganelle toolkit (Jin et al. Citation2020). Finally, compared to the plastome of the Taxillus chinensis (GenBank accession number: NC036306), it was annotated by Geneious Prime software (Kearse et al. Citation2012). For the fear of omitting annotations, we also manually adjusted annotations by comparing it with other plastomes of Loranthaceae that can be available on GenBank.
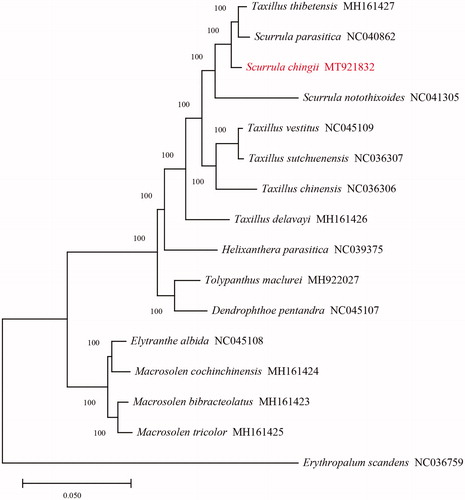
To confirm the phylogenetic position of the S. chingii, we downloaded 14 species plastomes of Loranthaceae from the GenBank and set the Erythropalum scandens (Olacaceae) (GenBank accession number: NC036759) as the outgroup to construct the phylogenetic tree. We used the MAFFT toolkit with default parameters to align the whole plastome sequences with one IR region (Katoh and Standley Citation2013). The maximum-likelihood (ML) tree was constructed using GTRCAT model with 1000 bootstrap iterations on the CIPRES portal (https://www.phylo.org) by RAxML-HPC2 toolkit (Miller et al. Citation2010).
The plastome of S. chingii (GenBank accession number: MT921832) is 122,764 bp in length, which consists of a large single-copy region (70,726 bp), a small single-copy region (6091 bp), and a pair of inverted repeat regions (22,974 bp). The GC content of the whole plastome is 37.2%. Meanwhile, a total of 109 genes were annotated, including 69 CDS (protein-coding genes), eight rRNAs, and 32 tRNAs. Among them, seven genes (rpl16, atpF, rpoC1, trnLUAA, petB, petD, and rpl2) have the single intron and three genes (rps12, ycf3, and clpP) have two introns. The special feature of the plastome, as same as the plastome of the tobacco, is that rps12 consists of three exons and its 5′ exon (5′-rps12) is located downstream from the other exons (3′-rps12) in IRb on the same strand, or downstream from the 3′-rpsl2 in IRa on the opposite strand. rpl2 gene straddled LSC/IRa border, and trnl-UAG gene straddled IRa/SSC and SSC/IRb border and compared to the plastome of Nicotiana tabacum (GenBank accession number: NC001879), the NADH dehydrogenase complex proteins (ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, and ndhK), four genes of ribosomal proteins (rpl2, rps15, rps16, and rpl32) and five tRNA genes (trnA, trnG, trnI, trnK, and trnV) are missing.
The topology of the phylogenetic tree shows that the S. chingii clustered with Scurrula parasitica and Taxillus thibetensis as a monophyletic group with a 100% bootstrap value (). The study corroborated the close phylogenetic relationship between Scurrula and Taxillus. The placement of T. thibetensis, however, conflicts with the former study by Liu et al. (Citation2018). We suspect the reason could be different DNA regions were used in the analysis and due to hemiparasitic plants do not wholly depend on their photosynthetic capacity, some genes of the plastome are lost. Similarly, the phenomenon of gene loss was also found in the other two hemiparasitic species, Taxillus chinensis and T. sutchuenensis (Li et al. Citation2017). Therefore, the complete chloroplast genome and phylogeny of hemiparasites from the Loranthaceae deserve further research.
Acknowledgements
We would like to thank the Molecular Biology Experiment Center, Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The plastome data of S. chingii (accession number is MT921832) using in this manuscript are deposited in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/), which can be searched after being examined and processed. We declare that the data should only be shared when not violating the protection of human subjects, or other valid ethical, privacy, or security concerns.
Additional information
Funding
References
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li J-L, Wang S, Yu J, Wang L, Zhou S-L. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48:72–78.
- Li Y, Zhou J-G, Chen X-L, Cui Y-X, Xu Z-C, Li Y-H, Song J-Y, Duan B-Z, Yao H. 2017. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species. Sci Rep. 7(1):12834.
- Liu B, Le C-T, Barrett RL, Nickrent DL, Chen Z-D, Li L-M, Vidal-Russell R. 2018. Historical biogeography of Loranthaceae (Santalales): diversification agrees with emergence of tropical forests and radiation of songbirds. Mol Phylogenet Evol. 124:199–212.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans, LA. p. 1−8.
- Press MC, Phoenix GK. 2005. Impacts of parasitic plants on natural communities. New Phytol. 166(3):737–751.
- Wang X-N, Zhang L. 2017. Species diversity and distribution of mistletoes and hosts in four different habitats in Xishuangbanna, Southwest China. J Yunnan Univ (Nat Sci). 39(4):701–711.