Abstract
Curculigo orchioides Gaertn. distributed in subtropical regions of Asia including southern China and India. The plant is used as a traditional medicine in China for the treatment of menorrhagia, osteoporosis, and other gynecological problems. The complete chloroplast genome was reported in this study using the Illumina NovaSeq platform. The whole genome of this species was 157,472 bp in length, with a total GC content of 37.44%. The large single copy (LSC) was 86,507 bp, the small single copy (SSC) was 16,867 bp, and both of the two inverted repeats (IRs) were 27,049 bp, respectively. A total of 132 unique genes were identified, among which are 86 protein-coding genes, 38 tRNA genes and 8 rRNA genes. The phylogenetic analysis revealed that C. orchioides was highly clustered with C. capitulata. Our study will provide useful fundamental data for further phylogenetic and evolutionary studies of C. orchioides.
Curculigo orchioides Gaertn. is a perennial herb of Amaryllidaceae, which grows widely in subtropical regions of Asia, such as southern China and India. The plant is used in the clinics of traditional Chinese and Indian medicine for many diseases such as osteoporosis and gynecological problems (Bafna and Mishra Citation2005; Jiao et al. Citation2009; Wang et al. Citation2012). The extracts of this plant contain a wide variety of phenolic glycosides, lignans, alkaloids, flavones, saponins, and other types of compounds (Valls et al. Citation2006; Zhou et al. Citation2020). The study also showed that the ethanol extracts of C. orchioides have the effect of strengthening the sexual behavior in male rats (Chauhan et al. Citation2007), and also have potential estrogenic activity in ovariectomized female albino rats (Vijayanarayana et al. Citation2007).
In recent years, with the increasing demand for C. orchioides, people have been digging and harvesting wild resources more immoderately. Moreover, because the rhizome of C. orchioides grows very slowly, and it has been threatened by habitat alternations, the resources of C. orchioides are significantly decreasing. For the above reasons, the medical effect and artificial cultivation of this plant have been intriguing the interests of researchers (Chen et al. Citation2017; Zhang et al. Citation2017; Zhou et al. Citation2020), while the complete chloroplast genome has not been sequenced. Considering the chloroplast DNA-based studies can provide invaluable data for studying genetic history and phylogeny, and can also provide important information in designing conservation and utilization strategies for the species, in this study, C. orchioides were collected from Laohuchong village, Qiubei county of Yunnan province (24°2773′ N, 103°8983′ E, 2228 m above sea level). A voucher specimen (YAB 202,007) was deposited at Yunnan Academy of Biodiversity, Southwest Forestry University, Yunnan, China. Then we sequenced, assembled and annotated the accurate chloroplast genome with the next-generation sequencing method, and the results will provide more useful information for phylogenetic and evolutionary research of this species.
For this study, the total genomic DNA of C. orchioides was extracted from fresh leaves according to the modified CTAB methods (Doyle and Doyle Citation1987). A genomic shotgun library with an insertion size of 341 bp was constructed, the libraries were sequenced on Illumina NovaSeq platform at Personalbio Biotech (Shanghai, China). The chloroplast genome was assembled using GetOrganelle software version 1.7.1 (Jin et al. Citation2020), and the assembled chloroplast genome was annotated through the online program CPGAVAS 2 (Shi et al. Citation2019) with C. capitulata chloroplast genome (GenBank accession number: MT610372) as a reference, and assisted with manual correction. The raw sequencing reads used in this study have been deposited in SRA (accession number: SRR12793630) and the annotated chloroplast genome sequence has been deposited into the GenBank (accession number: MW079480).
The complete chloroplast genome of C. orchioides was 157,472 bp and composed of two IRs of 27,049 bp each, which divide a large single copy (LSC) region of 86,507 bp and a small single copy (SSC) region of 16,867 bp, the average GC content was 37.44%, with IR regions (42.53%) higher than that in LSC (35.44%) and SSC regions (31.37%). The chloroplast genomes encoded 132 unique genes, including 86 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. A total of 86 SSR markers ranging from mononucleotide to pentanucleotide repeat motif were identified in the chloroplast genome of C. orchioides. The intron-exon structure analysis indicated that 17 genes have introns, among which atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC have one intron, while ycf3 and clpP have two introns.
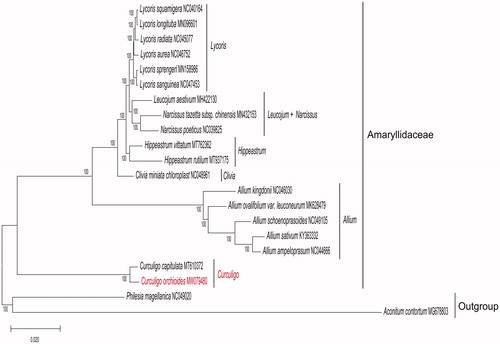
To determine the phylogenetic relationship of C. orchioides, based on complete chloroplast genomes of the other 18 species within the family Amaryllidaceae (), chloroplast genomes were downloaded from NCBI. All chloroplast genomes were aligned using the program MAFFT v7.471 (Rozewicki et al. Citation2019), and phylogenetic tree (maximum likelihood) constructed by Iqtree software version 1.6.12 (Minh et al. Citation2020) with 1000 bootstrap replicates, best-fitted model has been confirmed is TVM + F+R2 by ModelTest-NG (Darriba et al. Citation2020). The phylogenetic analysis revealed that C. orchioides closely clustered with C. capitulata. The study will provide essential data for future research on the phylogenetic and evolutionary relationship of C. orchioides and the family Amaryllidaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW079480. The associated BioProject, SRA and Bio-Sample numbers are PRJN667995, SRR12793630, and SAMN16393142 respectively.
Additional information
Funding
References
- Bafna AR, Mishra SH. 2005. In vitro antioxidant activity of methanol extract of rhizomes of Curculigo orhioides Gaern. Ars Pharm. 46(2):125–138.
- Chauhan NS, Rao CV, Dixit VK. 2007. Effect of Curculigo orchioides rhizomes on sexual behaviour of male rats. Fitoterapia. 78(7–8):530–534.
- Chen XL, Zuo AX, Deng ZT, Huang XY, Zhang XM, Geng CA, Li TZ, Chen JJ. 2017. New phenolic glycosides from Curculigo orchioides and their xanthine oxidase inhibitory activities. Fitoterapia. 122:144–149.
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jiao L, Cao DP, Qin LP, Han T, Zhang QY, Zhu Z, Yan F. 2009. Antiosteoporotic activity of phenolic compounds from Curculigo orchioides. Phytomedicine. 16(9):874–881.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Rozewicki J, Li SL, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10.
- Shi LC, Chen HM, Jiang M, Wang LQ, Xi W, Huang LF, Chang L. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Valls J, Richard T, Larronde F, Leblais V, Muller B, Delaunay JC, Monti JP, Ramawat KG, Mérillon JM. 2006. Two new benzylbenzoate glucosides from Curculigo orchioides. Fitoterapia. 77(6):416–419.
- Vijayanarayana K, Rodrigues RS, Chandrashekhar KS, Subrahmanyam EVS. 2007. Evaluation of estrogenic activity of alcoholic extract of rhizomes of Curculigo orchioides. J Ethnopharmacol. 114(2):241–245.
- Wang Y, Zhao L, Wang Y, Xu JL, Nie Y, Guo YH, Tong YT, Qin LP, Zhang QY. 2012. Curculigoside isolated from Curculigo orchioides prevents hydrogen peroxide-induced dysfunction and oxidative damage in calvarial osteoblasts. Acta Bioch Bioph Sin. 44(5):431–441.
- Wu XY, Li JZ, Guo JZ, Hou BY. 2012. Ameliorative effects of curculigoside from Curculigo orchioides Gaertn on learning and memory in aged rats. Molecules. 17(9):10108–10118.
- Zhang H, He G, Liu XG, Liang WB, Zeng BQ, Liu J, Ni SG, Zhang SL. 2017. Preliminary study on Curculigo orchiodes Gaertn artificial seeds. Mol Plant Breed. 15(8):3173–3178.
- Zhou F, Yao M, Wu Q, Wang SC, Wang W, Liu CX, Li B, Peng CY. 2020. Research progress on chemical constituents and pharmacological activities of Curculigo orchioides. Chin Tradit Herbal Drugs. 51(8):2238–2247.