?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The chloroplast genomic information was obtained from three wild Solanum and four hybrids by chloroplast genome sequencing. The chloroplast genomes of the seven samples comprise of a circular structure and sizes from 155,581 to 155,612 bp and composed of 130 genes. The genome structures of the two hybrids were identical, while the other two hybrids showed 2 bp differences in the LSC when compared with their maternal parent. The total sites of SNP and InDel were 39–344 and 54–90, respectively. With the exception of one hybrid with two additional sites, the other hybrids were identical to their maternal.
1. Introduction
Solanum L. is a rich genus of plants comprising ∼1400 diverse species, as one of the largest flowering plant genera (Bentolila and Stefanov Citation2012; Lim Citation2013). Solanum species are worldwide, including Antarctica, with wild resources found across southeast Asia, Africa, South America, and south China (Giacomin et al. Citation2013). Wild Solanum has several beneficial properties, including resistant and strong growth characteristics (Christodoulakis et al. Citation2009; Meyer et al. Citation2012; Rotino et al. Citation2014). In particular, the aubergine Solanum melongena harbors numerous genes that are beneficial for long-term breeding processes (Augustinos et al. Citation2016), which can be introduced to wild resource species through crossing to broaden the genetic basis as an important means of crop improvement and germplasm innovation. To date, S. melongena has been hybridized with Solanum aethiopicum, Solanum anthropophagoru, Solanum sysimbriifolium, and other closely related wild relatives of Leptostemonum, showing a significantly higher affinity than other species such as Solanum pecteinocarpum and Solanum aviculare (Poczai et al. Citation2008; Frary et al. Citation2011; Plazas et al. Citation2016).
Studies on the distant hybridization of Solanum have shown that the traits of hybrid offspring were more similar to those of wild resources with strong heterosis (Rotino et al. Citation2014; Kaushik et al. Citation2016). Although the growth and development advantages of distant hybrid offspring are known to be correlated with photosynthesis, most of the research on heterosis conducted to date has mainly focused on nuclear gene expression and regulation (Birchler et al. Citation2010; Meyer et al. Citation2010; Birchler et al. Citation2016; Jensen et al. Citation2018; Wang et al. Citation2018) and there is no information available as to whether chloroplasts play a role in Solanum heterosis. Heterosis is now widely used in the commercial production of many crop hybrids. For hybrid seed production, a controlled pollination system should be developed to prevent unnecessary self-pollination. Hence, male sterility plays an important role in heterosis utilization. It is of great theoretical and practical significance to explore and utilize the male-sterile new germplasm of plants so as to prevent an epidemic of diseases and insect pests in heterosis and promote yield improvements. Most Solanum interspecific hybrids are sterile (Afful et al. Citation2018), which provide precious materials to use and study. Further, most researchers think male sterility results from disturbed mitochondrial–nuclear interaction (Bentolila and Stefanov Citation2012; Liu et al. Citation2013), and some have reported the effects of chloroplast genome on plant male sterility (Liu Citation1983; He Citation2003).
The chloroplast is a plastid, which is a semi-autonomous plant organelle containing an independent genome (i.e. plastid DNA) with a length between 120 and 230 kb that encodes 100–130 proteins in higher plants. The transcription and translation of plastid genes have many conservative prokaryotic characteristics (Bock Citation2007; Alice Citation2011), and chloroplasts harbor the complete enzymes required for photosynthetic substrates and electron carriers in the thylakoid membranes. Chloroplast genome analyses have revealed two main modes of plastid inheritance in angiosperms: biparental inheritance and uniparental inheritance. Maternal inheritance is most common in angiosperms (Xiaohua et al. Citation2003; Jansen and Ruhlman Citation2012; Li et al. Citation2013), with only a few plastids being regularly or occasionally inherited in a biparental manner (Corriveau and Coleman Citation1988), whereas paternal plastids are more common in gymnosperms (Shiyi Citation1997), such as Medicago sativa (Diatchenko et al. Citation1996; Pellet et al. Citation1996), Daucus L (Iturbe-Ormaetxe et al. Citation2003), and Pharbitis (Hayashi et al. Citation2004). The cytoplasmic inheritance of Solanum tuberosum is maternal (Hu et al. Citation2008), which results from the decay of the male organelles themselves or the degradation of DNA, leading to the degradation and disappearance of plastids during the maturation process (Hagemann and Schröder Citation1989).
The chloroplast is an important photosynthetic organ in higher plants, and has a close relationship with major agricultural aspects, such as crop yield, tinea genetic, cytoplasmic male sterility, photosynthetic efficiency; thus, the deep study of higher plant chloroplast genetics is necessary. To determine the precise genetic pattern of plant chloroplasts is of great significance for phylogeny, biogeography, hybridization, and systematics.
Therefore, we investigated both the intraspecific and interspecific genetic patterns of chloroplast genomes among different hybrid combinations of Solanum and further explored the relationship between heterosis, male sterility, and chloroplast genomic effects. Specifically, we assembled the chloroplast genomes of three close self-crossing species of Solanum and their four interspecific hybrid F1 lines. These results can provide valuable baseline information on the genetic pattern of chloroplasts in the interspecific hybridization of Solanum.
2. Results
2.1. Genome assembly and genetic features
In total, 134,071 Mb of raw data were obtained from the seven species on the Illumina HiSeq4000 and Pabico platforms. From each species, 4317–6129 Mb of clean data were used to assemble the complete chloroplast genome of Solanum. The chloroplast genome comprises a double-stranded circular DNA sequence with a length of 155,581–155,612 bp. The chloroplast genome consists of a large single-copy (LSC) region of 86,160–86,196 bp, small single-copy (SSC) region of 18,501–18,562 bp, and two inverse repeat (IR) regions, IRA, and IRB, of 25,440–25,443 bp each. The GC contents of the seven chloroplast genomes were similar at 37.7–37.71%, which is in line with that previously reported for S. melongena (Ding et al. Citation2016).
The chloroplast genome structure of the hybrids S. melongena (177) × S. aethiopicum (y11) and S. aethiopicum (53) × S. aethiopicum (y11) was identical to that of their respective maternal genomes, whereas S. melongena (177) × S. torvum showed a 2-bp difference and S. aethiopicum (y11) × S. melongena (177) had a 4-bp, 1-bp, and 2-bp reduction in the LSC, IR, and SSC, respectively, relative to the maternal plant; all hybrids had GC content identical to that of the maternal plant ().
Table 1. Comparison of chloroplast sequences of the seven samples.
The Solanum chloroplast genome contains 130 (106 unique) functional genes, including 80 (71 unique plus ycf (#1, #2, #15) as hypotheticals) protein-coding genes, 42 (31 unique) tRNA genes, and eight (four unique) rRNA genes. Furthermore, 17 genes were found to be replicated in both IR regions, including six protein-coding genes and 11 RNA genes (four rRNAs and seven tRNAs; ). The LSC region contains 60 protein-coding and 27 tRNA genes, while the SSC region has 11 protein-coding genes and one tRNA gene ( and ).
Figure 1. Chloroplast reference genome of seven Solanum species. Genes shown inside the outer circle are transcribed counter clockwise and those outside are transcribed clockwise. Genes belonging to different functional groups are color-coded. The gray area in the inner circle indicates the GC content (see and for sample details).
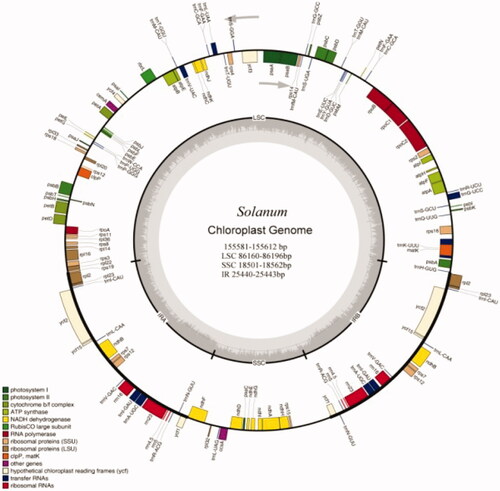
Table 2. List of genes encoded in the chloroplast genome of the seven Solanum species.
Table 3. SNP types of the chloroplast genomes of the seven Solanum species.
2.2. Comparative genome analysis
Using KU682719 (S. melongena plastid) as the reference, an analysis of the single nucleotide polymorphisms (SNPs) of the seven chloroplast genomes () showed that the hybrid S. aethiopicum (y11) × S. melongena (177) had one additional synonymous substitution in the coding sequence area and intergenic area, respectively. However, the other three hybrids were completely consistent with their maternal plants with respect to the quantity, type, and location of SNPs, with vast differences observed between different female parents. There was a total of 39 SNP sites in 177 (S. melongena) and its two hybrids, 76.9% of which were in the intergenic region. S. aethiopicum 53 and y11 had 339 and 342 SNPs, respectively, with few differences between them, and 71% of the SNP sites were in the intergenic region.
Using KU682719 (S. melongena plastid) as the reference, an analysis of the insertions/deletions (InDels) of the seven chloroplast genomes () showed that S. aethiopicum (y11) × S. melongena (177) has two additional loci in the intergenic region, and the other three hybrids were completely consistent with their maternal plants with regards to quantity, type, and location of InDels, with large differences among the different female parents. There were 54 InDel sites of 177 (S. melongena) and its two hybrids (177 as the maternal plant), 85.2% of which were in the intergenic region. S. aethiopicum 53 and y11 had 88–92 InDels, with minimal difference between them, and 63% of the InDel sites were in the intergenic region. The predominant InDel type was insertions, accounting for more than 63% of all InDels identified.
Table 4. InDel types of the chloroplast genomes of the seven Solanum species.
2.3. Phylogenetic analysis
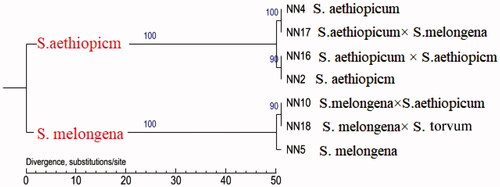
The assembled NN2 chloroplast genome was used as the reference genome for the construction of the phylogenetic tree based on the SNP data of the chloroplast genomes of the seven sequenced materials. As shown in , the phylogenetic tree was mainly divided into two branches: one branch made up of S. melongena and its hybrids S. melongena × S. aethiopicum and S. melongena × S. torvum, and the other made up of S. aethiopicum and its hybrids S. aethiopicum × S. melongena and S. aethiopicum × S. aethiopicum. This result indicates that the chloroplast of Solanum is maternally inherited.
3. Discussion
The cytoplasmic genetic patterns of more than 600 species of angiosperms have been identified to date (Nagata Citation2010). The most common methods currently used to identify the genetic pattern of chloroplasts are cytogenetics, use of fluorescent dyes (Quan and Yang Citation2003), molecular biology approaches (Testolin and Cipriani Citation1997; Xiaohua et al. Citation2003; Hansen et al. Citation2007; Zora and Pal Citation2007), and direct high-throughput sequencing. However, it is difficult to identify spermatozoa from the ultrastructure because of only a few morphological differences between male and female gametes, especially for plastids that are closely related. Fluorescence staining is a simple, sensitive, and rapid technique, which can directly detect plastids or mitochondria; however, it is difficult to distinguish between plastids and mitochondria with this approach. Molecular biology methods, including Southern hybridization or random primer amplification, can be directly used to determine the presence and transmission of plastids or mitochondria; however, these methods are hindered by the limited number of available probes or primers, making it difficult to determine the genetic status of organelles accurately and comprehensively. With the development of next-generation sequencing technology, a large amount of chloroplast sequence data can be obtained quickly and in a cost-effective manner, which is also the most direct method to determine the genetic mode of inheritance.
The genome structure, GC content, and annotated genes, in addition to the quantity, type, and location of SNPs and InDels of the hybrids are nearly the same as those in the maternal plant, which showed that the Solanum genus presents the typical pattern of matrilineal chloroplast inheritance. This has important implications for Solanum speciation because isolation and differentiation are the basic mechanisms of new species formation; however, there is no strict reproductive isolation between Solanum species, and the pollen has a certain interspecies affinity, which results in a wide variety of hybrid materials and local varieties that can be produced. Consequently, there is substantial debate regarding the evolutionary relationships and classifications of these intermediate hybrid materials and wild species. The present study confirmed that the Solanum genus shows the typical pattern of matrilineal chloroplast inheritance. Although different species could hybridize, they cannot form a new taxon.
Furthermore, the distant hybrids between Solanum species have excellent characteristics, including improved stress resistance (Collonnier et al. Citation2003; Iwamoto et al. Citation2007; Zhou et al. Citation2018), growth, and development (unpublished). However, hybrids share the same chloroplast system as their maternal counterparts. The hybrid formation process does not involve the recombination of chloroplast genomes of both parents, which means that heterosis has no relation with the chloroplast genome, but rather results from nuclear gene interactions that require further research.
In particular, CMS in plants is mainly caused by the nucleus of a species being replaced with a different cytoplasm, and many CMS materials are obtained through interspecific or intergeneric hybridization. In this study, three interspecific hybrids were sterile, which likely resulted from disturbed mitochondrial–nuclear interactions rather than the effects of the chloroplast genome. Our study shows that sterile F1s possess the same chloroplast genome similar to those of their maternal counterparts; therefore, the Solanum male sterility observed in this study is unrelated to the chloroplast genome.
4. Materials and methods
The self-cross-breeding materials of three Solanum species and the four hybrids were obtained from the Vegetable Research Institute seed bank, Guangxi Academy of Agricultural Science (28°N and 118°E) ().
Table 5. Basic information of the seven Solanum plants.
4.1. DNA extraction, genome sequencing, and assembly
Approximately 5 g of fresh leaves were harvested for cpDNA isolation, using an improved extraction method (McPherson et al. Citation2013). After DNA isolation, 1 μg of purified DNA was fragmented to construct short-insert libraries (insert size 430 bp) according to the manufacturer’s instructions (Illumina), followed by sequencing on the Illumina HiSeq 4000 (Borgström et al. Citation2011). The high molecular weight DNA was purified and used for PacBio library preparation, Blue Pippin size selection, and then sequenced on the Sequel Sequencer. Prior to assembly, Illumina raw reads were filtered. This filtering step was performed to remove the reads with adaptors, reads showing a quality score below 20 (Q < 20), reads containing a percentage of uncalled based (‘N’ characters) equal or greater than 10%, and duplicated sequences. The chloroplast genome was reconstructed using a combination of PacBio Sequel data and Illumina HiSeq data, and the following three steps were used to assemble the cp genomes (Richard Citation2008). First, the genome framework was assembled using both Illumina and PacBio data by SPAdes v3.10.1 (Antipov et al. Citation2016). Second, the assembly was verified, and the circle characteristics of the cp genomes were completed, filling any gaps. Third, clean reads were mapped to the assembled cp genome to correct the incorrect bases, and insertion and deletion were assessed.
Genome annotation
The chloroplast genes were annotated using the online DOGMA tool (Wyman et al. Citation2004), using default parameters to predict protein-coding genes, tRNA genes, and rRNA genes. A whole chloroplast genome Blast (Lobo Citation2012) search (E-value 10-5, minimal alignment length percentage
40%) was performed against five databases: KEGG (Kaneisha et al. Citation2004, Citation2006), COG (Tatusov et al. Citation1997, Citation2003), NR, Swiss-Pro (Magrane Citation2011), and GO (Ashburner et al. Citation2000). The circular Solanum chloroplast genome map was drawn using Organellar Genome DRAW v1.2 (Lohse et al. Citation2007).
The cp genome among 13 species was compared using the VISTA program. Genome, protein-coding gene, intron, and spacer sequence divergences were evaluated using DnaSP 5.10 after alignment. The genomic sequences were aligned using MAFFT v5, and were adjusted manually where necessary. For the protein-coding gene sequences, introns, and spacers, every gene or fragment was edited using the ClustalW multiple alignment option within the software BioEdit v7.0.9.0.
4.2. Comparative genome analysis
MUMmer and BLAST were used to conduct global alignment and local alignment between the sample sequence and the reference genome, determining potential SNPs. Subsequently, the SNPs were filtered out in the repeat regions as detected using the software BLAST, Repeat Masker, and TRF. Finally, SNPs were annotated based on the position and interaction between genes.
LASTZ software was also used to perform global alignment between each sample sequence and the reference genome. Subsequently, the alignment result was corrected by axt_correction, axtSort, and axtBest to determine potential InDels with lengths less than 50 bp. Finally, BWA and SAM tools were used to map the reads to InDel sequences and to filter out unreliable InDels.
4.3. Phylogenetic analysis
ClustalW was used to align the cpDNA sequences under default parameters (Larkin et al. Citation2007), and the alignment was checked manually. The maximum-likelihood (ML) methods were performed for genome-wide phylogenetic analyses using PhyML 3.0 (Guindon et al. Citation2010). Selection of the nucleotide substitution model was done using jModelTest 2.1.10 (Darriba et al. Citation2012) and Smart Model Selection in PhyML 3.0. The model GTR + G was selected for ML analyses with 1,000 bootstrap replicates to calculate the bootstrap values (BS) of the topology. The results were treated with iTOL 3.4.3 (Letunic and Bork Citation2016).
5. Conclusions
In this study, we used a high-throughput sequencing approach to characterize the chloroplast genome of Solanum, including three wild species and four hybrids to determine the mode of inheritance, extent of heterosis through hybridization, phylogenetic relationships, and the relationship between the male stile and chloroplast genome. We believe that our study makes a significant contribution to the literature because appropriate cross-breeding can be used to obtain hybrid varieties with excellent characteristics for crops, including improved growth and stress resistance, particularly for aubergine (S. melongena) as an important Solanum crop worldwide. Therefore, understanding the contribution of cytoplasmic inheritance and the chloroplast genome to these traits can aid in obtaining useful germplasm. Moreover, our results provide further information on chloroplast inheritance toward gaining a better fundamental understanding and facilitating further research on the interspecific heterosis, male sterility mechanism, and evolution of Solanum.
| Abbreviations | ||
| BS | = | bootstrap |
| CML | = | cytoplasmic male sterility |
| COG | = | clusters of orthologous groups |
| cpDNA | = | chloroplast DNA |
| GO | = | gene ontology |
| KEGG | = | Kyoto Encyclopedia of Genes and Genomes |
| InDel | = | insertion/deletions |
| LSC | = | large single-copy |
| NR | = | Non-Redundant Protein Database |
| SNP | = | short nucleotide polymorphism |
| tRNA | = | transfer RNA |
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The raw data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers: SAMN16746488, SAMN16746490, SAMN16746491, SAMN16746496, SAMN16746503, SAMN16746502, and SAMN16746504.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Afful NT, Nyadanu D, Akromah R, Amoatey HM, Annor C, Diawouh RG. 2018. Evaluation of crossability studies between selected eggplant accessions with wild relatives S. torvum, S. anguivi and S. aethopicum (Shum group). J Plant Breed Crop Sci. 10(1):1–12.
- Alice B. 2011. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155(4):1520–1532.
- Antipov D, Korobeynikov A, McLean JS, Pevzner PA. 2016. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 32(7):1009–1015.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. Nat Genet. 25(1):25–29.
- Augustinos AA, Petropoulos C, Karasoulou V, Bletsos F, Papasotiropoulos V. 2016. Assessing diversity among traditional Greek and foreign eggplant cultivars using molecular markers and morphometrical descriptors. Span J Agric Res. 14(4):e0710.
- Bentolila S, Stefanov S. 2012. A reevaluation of rice mitochondrial evolution based on the complete sequence of male-fertile and male-sterile mitochondrial genomes. Plant Physiol. 158(2):996–1017.
- Birchler JA, Johnson AF, Veitia RA. 2016. Kinetics genetics: incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Sci. 245:128–134.
- Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. 2010. Heterosis. Plant Cell. 22(7):2105–2112.
- Bock R. 2007. Structure, function, and inheritance of plastid genomes. In: Bock R. (eds) Cell and Molecular Biology of Plastids. Topics in Current Genetics, vol 19. Springer, Berlin, Heidelberg
- Borgström E, Lundin S, Lundeberg J. 2011. Large scale library generation for high throughput sequencing. PLoS One. 6(4):e19119.
- Christodoulakis NS, Lampri PN, Fasseas C. 2009. Structural and cytochemical investigation of the leaf of silverleaf nightshade (Solanum elaeagnifolium), a drought-resistant alien weed of the Greek flora. Aust J Bot. 57(5):432–438.
- Collonnier C, Fock I, Daunay M-C, Servaes A, Vedel F, Siljak-Yakovlev S, Souvannavong V, Sihachakr D. 2003. Somatic hybrids between Solanum melongena and S. sisymbrifolium, as a useful source of resistance against bacterial and fungal wilts. Plant Sci. 164(5):849–861.
- Corriveau JL, Coleman AW. 1988. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot. 75(10):1443–1458.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 93(12):6025–6030.
- Ding QX, Liu J, Gao LZ. 2016. The complete chloroplast genome of eggplant (Solanum melongena L). Mitochondrial DNA Part B-Res. 1(1):843–844.
- Frary A, Tümbilen Y, Doganlar S, Daunay M-C. 2011. Application of EST-SSRs to examine genetic diversity in eggplant and its close relatives. Turk J Biol. 35(2):125–136.
- Giacomin LL, Bohs L, Stehmann JR. 2013. Two new species from the Brevantherum clade of Solanum (solanaceae) from Eastern Brazil. J Bot Res Inst Texas. 7(1)
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hagemann R, Schröder M-B. 1989. The cytological basis of the plastid inheritance in angiosperms. Protoplasma. 152(2–3):57–64.
- Hansen AK, Escobar LK, Gilbert LE, Jansen RK. 2007. Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogenetic studies. Am J Bot. 94(1):42–46.
- Hayashi H, Huang P, Takada S, Obinata M, Inoue K, Shibuya M, Ebizuka Y. 2004. Differential expression of three oxidosqualene cyclase mRNAs in Glycyrrhiza glabra. Biol Pharm Bull. 27(7):1086–1092.
- He G. 2003. A common sequence difference between cytoplasmic male sterile lines and their maintainer lines existing in rice (Oryza sativa L.) chloroplast tRNA-Leu gene region. Euphytica. 131(3):269–274.
- Hu YC, Zhang Q, Rao GY, Sodmergen. 2008. Occurrence of plastids in the sperm cells of Caprifoliaceae: biparental plastid inheritance in angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol 49:958–968.
- Iturbe-Ormaetxe I, Haralampidis K, Papadopoulou K, Osbourn AE. 2003. Molecular cloning and characterization of triterpene synthases from Medicago truncatula and Lotus japonicus. Plant Mol Biol. 51(5):731–743.
- Iwamoto Y, Hirai M, Ohmido N, Fukui K, Ezura H. 2007. Fertile somatic hybrids between Solanum integrifolium and S. sanitwongsei (syn. S. kurzii) as candidates for bacterial wilt-resistant rootstock of eggplant. Plant Biotechnol. 24(2):179–184.
- Jansen RK, Ruhlman TA. 2012. Plastid genomes of seed plants. In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Advances in photosynthesis and respiration (including bioenergy and related processes), Vol. 35, pp. 103-126. Dordrecht (The Netherlands): Springer.
- Jensen C, Ørsted M, Kristensen TN. 2018. Effects of genetic distance on heterosis in a Drosophila melanogaster model system. Genetica. 146(4–5):345–359.
- Kaneisha M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M, et al. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354–D357.
- Kaneisha M, Goto S, Kawashima S, Okuna Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32:D277–D280.
- Kaushik P, Prohens J, Vilanova S, Gramazio P, Plazas M. 2016. Phenotyping of eggplant wild relatives and interspecific hybrids with conventional and phenomics descriptors provides insight for their potential utilization in breeding. Front Plant Sci. 7:677.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Li D, Qi X, Li X, Li L, Zhong C, Huang H. 2013. Maternal inheritance of mitochondrial genomes and complex inheritance of chloroplast genomes in Actinidia Lind.: evidences from interspecific crosses. Mol Genet Genomics. 288(3–4):101–110.
- Lim TK. 2013. Solanum sessiliflorum.
- Liu C, Ma N, Wang P-Y, Fu N, Shen H-L. 2013. Transcriptome sequencing and de novo analysis of a cytoplasmic male sterile line and its near-isogenic restorer line in chili pepper (Capsicum annuum L.). PLoS One. 8(6):e65209.
- Liu Y. 1983. Chloroplast DNA and cytoplasmic male sterility. Theor Appl Genet. 10(2):114–122.
- Lobo I. 2012. Basic Local Alignment Search Tool (BLAST). J Mol Biol. 215(3):403–410.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Magrane M. 2011. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011:bar009.
- McPherson H, van der Merwe M, Delaney SK, Edwards MA, Henry RJ, McIntosh E, Rymer PD, Milner ML, Siow J, Rossetto M. 2013. Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol. 13(1):8.
- Meyer RC, Kusterer B, Lisec J, Steinfath M, Becher M, Scharr H, Melchinger AE, Selbig J, Schurr U, Willmitzer L, et al. 2010. QTL analysis of early stage heterosis for biomass in Arabidopsis. Theor Appl Genet. 120(2):227–237.
- Meyer RS, Karol KG, Little DP, Nee MH, Litt A. 2012. Phylogeographic relationships among Asian eggplants and new perspectives on eggplant domestication. Mol Phylogenet Evol. 63(3):685–701.
- Nagata N. 2010. Mechanisms for independent cytoplasmic inheritance of mitochondria and plastids in angiosperms. J Plant Res. 123(2):193–199.
- Pellet DM, Papernik LA, Kochian LV. 1996. Multiple aluminum-resistance mechanisms in wheat (roles of root apical phosphate and malate exudation). Plant Physiol. 112(2):591–597.
- Plazas M, Vilanova S, Gramazio P, Rodríguez-Burruezo A, Fita A, Herraiz FJ, Ranil R, Fonseka R, Niran L, Fonseka H, et al. 2016. Interspecific hybridization between eggplant and wild relatives from different genepools. J Amer Soc Hort Sci. 141(1):34–44.
- Poczai P, Taller J, Szabo I. 2008. Analysis of phylogenetic relationships in the genus Solanum (Solanaceae) as revealed by RAPD markers. Plant Syst Evol. 275(1–2):59–67.
- Quan Z, Yang L. 2003. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 44(9):941–951.
- Richard C. 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36(19):e122.
- Rotino GL, Sala T, Toppino L. 2014. Eggplant. Alien Gene Transfer in Crop Plants. 2(7):381–409.
- Shiyi h. 1997. Cytological study of plastid inheritance in angiosperms. Acta Botanica Sinica. (4):363–371.
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 4(1):41.
- Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science. 278(5338):631–637.
- Testolin R, Cipriani G. 1997. Paternal inheritance of chloroplast DNA and maternal inheritance of mitochondrial DNA in the genus Actinidia. Theor Appl Genet. 94(6–7):897–903.
- Wang Y, Zhang K, Sun L, Han X, Fan S, Li X, Qu Y, Yao D, Wang P, Zhang J, et al. 2018. Study on the relationship between genetic variation of DNA methylation and heterosis in soybean leaves. Euphytica. 214(5):85.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xiaohua F, Fang Z, Naihu W, Shiyi H. 2003. Plastid inheritance in sweet potato as revealed by DNA restriction fingerprinting. J Plant. 45(1):73–75.
- Zhou X, Bao S, Liu J, Yang Y, Zhuang Y. 2018. Production and characterization of an amphidiploid derived from interspecific hybridization between Solanum melongena L. and Solanum aculeatissimum Jacq. Sci Hortic. 230:102–106.
- Zora S, Pal M. 2007. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. PNAS. 104(17):7003.