Abstract
The mitochondrial genome of one Rana pseudo-rana species Rana sangzhiensis Shen was sequenced and annotated. The mitogenome is 19,207 bp in length, containing 37 typical genes. The A + T content of the whole mitogenome is 56.6%. All of the protein-condoning genes (PCGs) started with ATG and stopped with TGA. The tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, andND5 are located in the circular mitochondrial L chain. The phylogeny tree is monophyletic among 14 related Rana species. The R. sangzhiensis Shen cluster was more closely related to R. amurensis Boulenger and R. kunyuensis Lu, Y.-Y., and P.-P. Li. This mitochondrial genome can be used for further analyses of Ranidae mitochondrial comparative genomics to improve the understanding of diverse Ranidae species.
The Rana sangzhiensis as a kind of small and medium-sized frog belonging to Rana pseudo-rana, Ranidae. The R. sangzhiensis Shen is distributed in Chenzhou, Yongzhou, Hengyang, Loudi, Huaihua and Xiangxi City, Hunan province, China. The R. sangzhiensis Shen generally lives in stream migration water and the surrounding grass with the mountain altitude of 800–1000 meters above sea level, and comes out to hunt for food in the day, and in the night only to mate in the breeding season (Xiang-Rong et al. Citation2008).
A male adult of R. sangzhiensis Shen was collected from Xingning Town (Zixing, Chenzhou City, Hunan Province, China) (113°43.0′E, 27°99.9′N), in October 2019. The specimens (No. Zg-zx) were stored at −80 °C in our laboratory (College of Animal Science and Technology, Hunan Agriculture University). Total genomic DNA of the R. sangzhiensis Shen was extracted from fertilized eggs using the Column Animal DNA out (Beijing Tiandz, Beijin, China) following the kit’s instructions. Then, the mitochondrial genome (mitogenome) of R. sangzhiensis Shen was sequenced by Illumina sequencing platform (Wuhan Biowefind biotechnology Co., Ltd, Wuhan, China). The reads were assembled and annotated using Generous Prime (v2019.1.3.). All tRNA genes were identified by ARWEN v1.2 (Laslett and Canbäck Citation2008). The annotated sequences of mitogenome were submitted to GenBank with the accession number MT782121. All protein-condoning genes (PCGs) were aligned using MAFFT algorithm in the TranslatorX (Katoh et al. Citation2019), and then poorly aligned results were removed by Gblocks 9.1 b (Castresana Citation2000; Abascal et al. Citation2010).
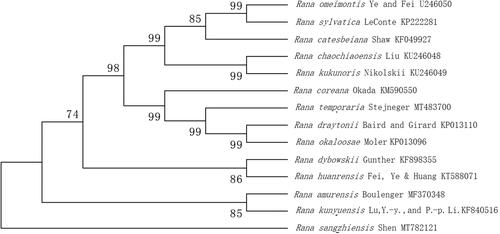
Mitogenome of R. sangzhiensis Shen has 19,207 bp in length, including 37 typical genes (13 PCGs, 22 tRNA genes and 2 rRNA genes). The overall nucleotide composition of the mitogenome is 27.3% for A, 28.5% for C, 14.7% for G and 29.3% for T. The A + T content of genes is 56.6%. All of PCGs started with ATG and stopped with TGA. The tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and ND5 are located in the circular mitochondrial L chain. The length of 22 tRNA is range from 67 bp (tRNA-Ser) to 74 bp (tRNA-Asn). Genes of 16S rRNA and 12S rRNA are 1576 bp and 930 bp, respectively. To validate the phylogenetic position of R. sangzhiensis Shen, we perform multiple sequence alignment by MEGA 6.0 (Tamura et al. Citation2013) and construct a maximum likelihood tree based on the complete mitochondrial genome sequences of 16 species in Ranidae. The phylogenetic tree () revealed that 14 Rana species constituted five subclades, the R. sangzhiensis Shen clustered in one clade, the R. sangzhiensis Shen cluster was more closely related to R. amurensis Boulenger and R. kunyuensis Lu, Y.-Y. and P.-P. Li.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT782121. The associated BioProject, SRA, and Bio-Sample numbers are 20TD07, PRJNA678467, and Zg-zx respectively.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38(Web Server issue):W7–W13.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Xiang-Rong He, Xiao-Qing Wang, Wu-Jiang Fan, Xian-Wen Zhou. 2008. Analysis of biological characters and nutritional components of Rana sangzhiensis. J Hunan Agric Univ (Nat Sci). 8.34(4):482–484.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.