Abstract
We report the complete chloroplast genome of the MED1 strain of Nephroselmis pyriformis from the Eastern Mediterranean Sea. At 111,026 bp, this genome is smaller and more compact than those of Nephroselmis olivacea and Nephroselmis astigmatica, and in contrast to the latter taxa, its inverted repeat contains no complete protein-coding genes. It encodes 3 rRNAs, 33 tRNAs and 94 proteins. Maximum likelihood analysis of a concatenated set of chloroplast genes from green algae belonging to deep-diverging lineages positioned the three Nephroselmis species in a strongly supported clade in which N. pyriformis is sister to N. astigmatica.
Representing a basal lineage of the Chlorophyta (Nephroselmidophyceae), the green algal genus Nephroselmis comprises 13 taxonomically accepted species of unicellular flagellates with a reniform shape and two unequal flagella (Leliaert et al. Citation2016; Guiry and Guiry Citation2020). Except for N. olivacea, which is freshwater, all Nephroselmis species inhabit marine or brackish waters. Nephroselmis pyriformis (N. Carter) Ettl 1982 (etymologically, ‘pear-shape’) can be distinguished based on the number and shape of its body scales (Yamaguchi et al. Citation2011). Known as a cosmopolitan species, N. pyriformis was first observed in the Eastern Mediterranean Sea in 2015 (Eker-Develi Citation2015). This identification, which was based on light microscopy, was later confirmed by Scanning Electron Microscopy and molecular barcoding of its partial SSU gene (GenBank: MN559709) (Konucu et al., Citation2019). A separate strain of N. pyriformis, strain MED1, was isolated in February 2016 from samples of the Eastern Mediterranean Sea taken near Erdemli (Turkey) (36°36′N, 34°19′E). In the present study, we report the chloroplast genome sequence of this strain and compare it with the genomes of the two other Nephroselmis species that are currently available in public databases: N. olivacea (GenBank: AF137379, 200,799 bp) (Turmel et al. Citation1999) and N. astigmatica (GenBank: KJ746600, 125,042 bp) (Lemieux et al. Citation2014).
The N. pyriformis MED1 strain, together with frozen biomass and DNA samples of this strain, are being kept in the Department of Biotechnology at the University of Mersin. DNA sequencing was performed by the Beijing Genomics Institute (Shenzhen, China) on the DNBSEQ platform. A total of ca. 40 million 150-bp paired-end reads were generated and assembled using SPAdes 3.14.0 (Bankevich et al. Citation2012). Following the identification of chloroplast contigs, the chloroplast genome sequence was completed using Consed (Gordon and Green Citation2013). Genes were identified as previously described (Turmel et al. Citation2017).
The N. pyriformis genome (GenBank: MW077730) contains 128 conserved genes that encode 91 proteins, 3 rRNAs, 33 tRNAs and a rnpB gene, and also displays 3 additional open reading frames (120, 160 and 186 amino-acid long). At 111,026 bp, it is smaller and more compact than the N. olivacea (200,799 bp) and N. astigmatica (125,042 bp) chloroplast genomes. Although gene content is well conserved among the three Nephroselmis chloroplast genomes, gene order is substantially scrambled. The N. pyriformis and N. olivacea chloroplast genomes are the most similar with respect to gene content, as both genomes share five of the seven genes that were previously found to be missing in N. astigmatica (accD, cemA, ftsI, ftsW, and rnpB) and as in the latter species, rne and tilS are lacking in N. pyriformis. The Long Single Copy (LSC) is 78,305 bp long, contains 74 protein-coding genes, a single non conserved ORF (ORF186a), rnpB and 25 tRNA. The Short Single Copy (SSC) is 19,693 bp long, contains 17 protein-coding genes, 2 non conserved ORFs (ORF120b and ORF160b) and 2 tRNA. The 6514-bp inverted repeat (IR) of N. pyriformis encodes 3 rRNA and 6 tRNA genes but no complete protein-coding gene, while the IRs of N. olivacea (46.1 kb) and N. astigmatica (13.7 kb) display several protein-coding genes in addition to rRNA and tRNA genes. As observed for N. olivacea, the N. pyriformis genome contains no introns. The putative protein encoded by ftsH is 3600 amino acid (aa) long in N. pyriformis, a size close to the corresponding N. olivacea sequence (3742 aa, GenBank: AAD54848) but much smaller than that of N. astigmatica (5,242 aa, GenBank: AID67672). The reverse situation was observed for ycf1, where the putative protein of N. astigmatica (AID67738, 1616 aa) is much longer than those of N. pyriformis (1074 aa) and N. olivacea (AAD54900, 956 aa). The putative proteins encoded by ycf20 were found to be poorly conserved among the three Nephroselmis species (showing less than 35% sequence identity in pairwise Clustal Omega (Sievers et al. Citation2011) alignments), supporting the view that this gene could be a pseudo-gene in Nephroselmis (Lemieux et al. Citation2014).
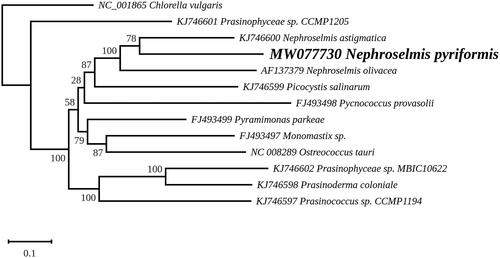
A maximum likelihood phylogeny was inferred using RAxML version 8.0 (Stamatakis 2014) and the set of 71 genes that Lemieux et al. (Citation2014) selected for their study of prasinophytes. Concatenated nucleotide sequences were aligned with MAFFT 7 (Katoh and Standley Citation2013) and variable regions in these alignments were trimmed with trimAl (Capella-Gutierrez et al. Citation2009). The phylogenic analysis was performed under the GTR + I + G model using Chlorella vulgaris (GenBank: NC_001865) as outgroup, with the best tree out of 100 being computed for 1,000 bootstrap replicates. The inferred tree identified the three Nephroselmis species within a strongly supported clade in which N. pyriformis is sister to N. astigmatica (), a result that is in agreement with the 18S rRNA phylogeny reported by Lubiana et al. (Citation2017). The presence of a gene-rich chloroplast genome in the basal N. olivacea taxa indicates that this feature is an ancestral characteristic of the genus and that several genes were lost in the marine lineage leading to N. astigmatica.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW077730. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA680225, SRR13108214, and SAMN16872557 respectively. The genome sequence data are also available on Zenodo with the following link: http://doi.org/10.5281/zenodo.4133421.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Eker-Develi E. 2015. First record of chlorophyte Nephroselmis pyriformis from the north-eastern Mediterranean Sea coast. Mar Biodivers Rec. 8:1–4.
- Guiry MD, Guiry GM. 2020. AlgaeBase. Galway: National University of Ireland; [accessed 2020 Oct 14]. http://www.algaebase.org.
- Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics. 29(22):2936–2937.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Konucu M, Tekdal D, Eker-Develi E. 2019. Kuzeydoğu Akdeniz Kıyı Sularında Tespit Edilen Prasinofit Nephroselmis pyriformis ve Diyatom Trieres mobiliensis Türlerinin FE SEM ve Moleküler Analiz ile Doğrulanması/Verification of The Prasinophyte Nephroselmis pyriformis and The Diatom Trieres mobiliensis by FE SEM and Molecular Analysis in The Northeastern Mediterranean Sea. Proceedings of the 2. International Mersin Symposium, 23–25 May 2019, Mersin, 178–193.
- Leliaert F, Tronholm A, Lemieux C, Turmel M, DePriest MS, Bhattacharya D, Karol KG, Fredericq S, Zechman FW, Lopez-Bautista JM. 2016. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci Rep. 6:25367.
- Lemieux C, Otis C, Turmel M. 2014. Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genom. 15(1):857.
- Lubiana KMF, Gianesella SMF, Saldanha-Corrêa FMP, Oliveira MC. 2017. Nephroselmis viridis (Nephroselmidophyceae, Chlorophyta), a new record for the Atlantic Ocean based on molecular phylogeny and ultrastructure. Mar Biodivers Rec. 10(1):5.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li R, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.
- Turmel M, Otis C, Lemieux C. 1999. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc Natl Acad Sci USA 96: 10248–10253.
- Turmel M, Otis C, Lemieux C. 2017. Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci Rep. 7(1):994.
- Yamaguchi H, Suda S, Nakayama T, Pienaar RN, Chihara M, Inouye I. 2011. Taxonomy of Nephroselmis viridis sp. nov. (Nephroselmidophyceae, Chlorophyta), a sister marine species to freshwater N. olivacea. J Plant Res. 124(1):49–62.