Abstract
Here we report the complete mitochondrial genome of the aphid species Mollitrichosiphum tenuicorpus. The M. tenuicorpus mitogenome is 15,727 bp in length and comprising 37 genes typically present in insect mitogenomes, a control region, and a unique repeat region. All protein-coding genes (PCGs) terminate with TAA or TAG except for cox1, which is terminated with T—. The secondary structure of trnS (AGN) loses the dihydrouridine (DHU) arm, but all the other transfer RNAs show the typical clover-leaf secondary structure. The repeat region between trnE and trnF is 458 bp, with a 217-bp repeat unit repeating 2.11 times. Phylogenetic analysis of the M. tenuicorpus mitogenome using the maximum-likelihood optimality criterion places it in a strongly supported sister position to Eutrichosiphum pasaniae. These data show that mitogenome sequences could be useful in resolving phylogenetic relationships of the Greenideinae.
Keywords:
The aphid species Mollitrichosiphum tenuicorpus (Okajima, 1908) (Aphididae: Greenideinae: Greenideini) is widely distributed in southeastern Asia. It is monoecious and holocyclic, mainly feeding on young shoots of the plants of Castanopsis, Litsea, and Lithocarpus (Blackman and Eastop Citation2020). To date, five complete mitochondrial genomes of Greenideinae species have been reported (Wang et al. Citation2014; Chen et al. Citation2019, Citation2020; Li et al. Citation2020; Liu et al. Citation2020). In this study, we sequenced and annotated the mitochondrial genome of M. tenuicorpus. The aphid specimens were collected on Lithocarpus glaber from Lishui City, Zhejiang, China (27.6909°N, 119.6352°E) and deposited in the National Zoological Museum of China, Institute of Zoology, Chinese Academy of Sciences, Beijing, China (NZMC no. 38234, Jing Chen, [email protected]). The sequencing was performed on an Illumina platform. The mitogenome was assembled using SPAdes version 3.10.1 (Bankevich et al. Citation2012) and the annotation was conducted using MITOS version 2 WebServer (Bernt et al. Citation2013), followed by manual adjustments.
The mitochondrial genome of M. tenuicorpus is 15,727 bp long, with an A + T content of 84.5% (GenBank accession number MW123009). The mitogenome contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), a control region, and a special non-coding repeat region. The gene order is identical to the inferred ancestral arrangement of insects (Clary and Wolstenholme Citation1985). All PCGs are initiated by ATN codons. Twelve PCGs are terminated with TAA or TAG codons, whereas cox1 uses the incomplete T— as the stop codon. The tRNA genes range from 62 to 73 bp in length. All tRNAs except for trnS (AGN) display the typical clover-leaf secondary structure. The dihydrouridine (DHU) arm is lost in the secondary structure of trnS (AGN). The rrnL and rrnS genes are located on the minority strand. The rrnL gene is 1271 bp long, with an A + T content of 85.7%. The rrnS gene is 769 bp in length, with an A + T content of 83.2%. The control region resides between rrnS and trnI. It is 680 bp long with an A + T content of 90.5%, including an AT-rich zone, a poly-thymidine stretch, and a stem-loop region. In the mitogenome of M. tenuicorpus, the repeat region located between trnE and trnF is 458 bp in length with an A + T content of 87.2%. Within this non-coding repeat region, a 217-bp repeat unit repeats 2.11 times. Up to now, this aphid-specific repeat region has been found in all reported mitogenomes of Greenideinae species except for Cervaphis quercus Takahashi, 1918.
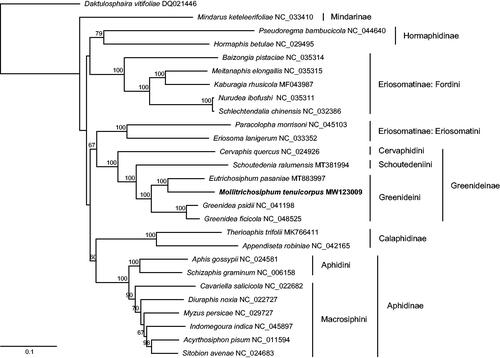
We built a maximum-likelihood tree of aphids using the whole mitogenome sequences of M. tenuicorpus and 26 other aphid species. The phylogenetic analysis was performed using RAxML version 8.2.10 (Stamatakis Citation2014). All subfamilies represented by more than one aphid species were recovered as monophyletic except for Eriosomatinae. The monophyly of Greenideinae and Greenideini was well supported with high bootstrap values. The tribe Cervaphidini was sister to Schoutedeniini + Greenideini, which is consistent with a previous phylogenetic study of Greenideinae based on multiple nuclear and mitochondrial genes (Liu et al. Citation2015). Within the clade of Greenideini, M. tenuicorpus and Eutrichosiphum pasaniae formed a sister group with strong support ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MW123009. The associated BioProject, SRA and Bio-Sample numbers are PRJNA681930, SRX9616531 and SAMN16974884, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Blackman RL, Eastop VF. 2020. Aphids on the world’s plants. [accessed 2020 Oct 16]. http://www.aphidsonworldsplants.info/.
- Chen J, Jiang L, Zhang X, Qiao G. 2020. The complete mitochondrial genome of Schoutedenia ralumensis Rübsaamen, 1905 (Hemiptera: Aphididae: Greenideinae). Mitochondrial DNA Part B. 5(3):2217–2218.
- Chen J, Wang Y, Qin M, Jiang LY, Qiao GX. 2019. The mitochondrial genome of Greenidea psidii van der Goot (Hemiptera: Aphididae: Greenideinae) and comparisons with other Aphididae aphids. Int J Biol Macromol. 122:824–832.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Li C, Jiang L, Zhang X, Chen J, Qiao G. 2020. The complete mitochondrial genome of Eutrichosiphum pasaniae (Okajima, 1908) (Hemiptera: Aphididae: Greenideinae). Mitochondrial DNA Part B. 5(3):3668–3669.
- Liu Q, Zhang H, Deng J, Lin X, Hang X. 2020. The complete mitochondrial genome of Greenidea ficicola (Hemiptera: Aphididae: Greenideinae), a pest of Ficus. Mitochondrial DNA Part B. 5(1):254–256.
- Liu QH, Chen J, Huang XL, Jiang LY, Qiao GX. 2015. Ancient association with Fagaceae in the aphid tribe Greenideini (Hemiptera: Aphididae: Greenideinae). Syst Entomol. 40(1):230–241.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang Y, Huang XL, Qiao GX. 2014. The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae). Insect Sci. 21(3):278–290.